For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STDGPDGQVGPAQRIAAGYDVDGQALQLGSVVIDGAADPAAQIRIPLATVNRHGLVAGATGTGKTKTLQL IAEQLSAAGVSVLMADVKGDLSGLSRAGQTDDKLAARAEETGDAWTPTAFPVEFLSLGAGGVGVPVRATI SSFGPILLAKVLGLNATQESTLGLIFHWADQRELPLLDLKDLRAVITHLISDEGKAELKSLGAVSPTTAG VILRALVNLEAEGADSFFGEPELRPDDLLRVDGQGRGIISLLELGSQSLRPVLFSTFLMWVLADLFTYLP EVGDLDKPKLVFFFDEAHLLFSDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDLPKDVLSQLGARIQH ALRAFTPDDHKALTKTVRTYPKTDVYDLESALTSLGIGEAVVTVLSEKGVPTPVAWTRMRAPRSLMGAIG SEAIRAVAQASPLQAVYGQTIDRESAHEMLSAKYAPATEAAEPPATAPQPPRGKYDPLPWPQDFDLPPMP TPVEPQGPPLWEEVLKNPTVKSAINTAAREFTRSIFGTGRRRRK*
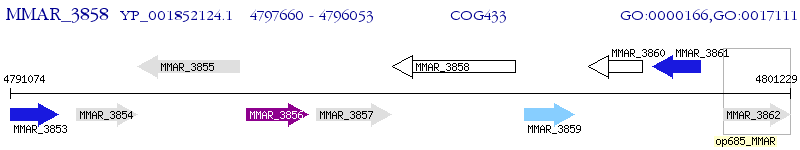
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3858 | - | - | 100% (535) | hypothetical protein MMAR_3858 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2538c | - | 0.0 | 87.50% (528) | hypothetical protein Mb2538c |
| M. gilvum PYR-GCK | Mflv_2544 | - | 0.0 | 74.21% (539) | protein of unknown function DUF853, NPT hydrolase putative |
| M. tuberculosis H37Rv | Rv2510c | - | 0.0 | 87.50% (528) | hypothetical protein Rv2510c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1534 | - | 0.0 | 69.51% (515) | hypothetical protein MAB_1534 |
| M. avium 104 | MAV_1665 | - | 0.0 | 72.83% (530) | ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_4723 | - | 0.0 | 75.71% (527) | hypothetical protein MSMEG_4723 |
| M. thermoresistible (build 8) | TH_1382 | - | 0.0 | 75.56% (532) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4099 | - | 0.0 | 76.70% (528) | protein of unknown function DUF853, NPT hydrolase putative |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2538c|M.bovis_AF2122/97 MGTESAAGGPGGPAQRIAAGYTVEGQALQLGTVVVDG----EPDPSAQIR
Rv2510c|M.tuberculosis_H37Rv MGTESAAGGPGGPAQRIAAGYTVEGQALQLGTVVVDG----EPDPSAQIR
MMAR_3858|M.marinum_M MSTDGPDGQVG-PAQRIAAGYDVDGQALQLGSVVIDG----AADPAAQIR
Mflv_2544|M.gilvum_PYR-GCK MTTD--PSG--TPAQQIAAGYAVEGAALELGAVVVDG----VCDPAARVR
Mvan_4099|M.vanbaalenii_PYR-1 MTAE--LSG--TPAQQIAAGYAVEGAALDLGSVVVDG----VCDPSARVR
MSMEG_4723|M.smegmatis_MC2_155 MTTE--STA--TPAQQIAAGYAVEGQALELGTVVVDG----AVDPTAQVR
TH_1382|M.thermoresistible__bu MTSG--STA--NPAQDIAAGYAVSGAALQLGSVMVDGPEGPVTDPDAQVR
MAB_1534|M.abscessus_ATCC_1997 MAEQ--TTAGATPAQQIAAGYATTGQALELGSVVVDG----TVDPAARIR
MAV_1665|M.avium_104 MSTD----SAATPATRIADGYAVAGQALELGTVVIDG----AADPSAQIR
* ** ** ** . * **:**:*::** ** *::*
Mb2538c|M.bovis_AF2122/97 IPLATVNRHGLVAGATGTGKTKTLQLIAEQLSAAGVAVLMADVKGDLSGL
Rv2510c|M.tuberculosis_H37Rv IPLATVNRHGLVAGATGTGKTKTLQLIAEQLSAAGVAVLMADVKGDLSGL
MMAR_3858|M.marinum_M IPLATVNRHGLVAGATGTGKTKTLQLIAEQLSAAGVSVLMADVKGDLSGL
Mflv_2544|M.gilvum_PYR-GCK IPLSTLNRHGLVAGATGTGKTKSLQVVAEQLSAAGVPVVMADVKGDLSGL
Mvan_4099|M.vanbaalenii_PYR-1 IPLATLNRHGLVAGATGTGKTKSLQVLAEQLSAAGVPVVMADVKGDLSGL
MSMEG_4723|M.smegmatis_MC2_155 IPLATVNRHGLIAGATGTGKTKSLQVLAEQLSAAGVPVLMADVKGDLSGL
TH_1382|M.thermoresistible__bu IPMATVNRHGLIAGATGTGKTKSLQLMAEQLSAAGVPVVMADIKGDLSGL
MAB_1534|M.abscessus_ATCC_1997 IPLATLNRHGLIAGATGTGKTKTLQVIAEQLSAAGVPVVMADVKGDLSGI
MAV_1665|M.avium_104 IPLATVNRHGLVAGATGTGKTKTLQLIAEQLSAAGVPVLMADVKGDLSGL
**::*:*****:**********:**::*********.*:***:******:
Mb2538c|M.bovis_AF2122/97 ARPGEAADKTAARAKDTGDDWVPTAFPVEFLSLGASGVGVPVRATISSFG
Rv2510c|M.tuberculosis_H37Rv ARPGEAADKTAARAKDTGDDWVPTAFPVEFLSLGASGVGVPVRATISSFG
MMAR_3858|M.marinum_M SRAGQTDDKLAARAEETGDAWTPTAFPVEFLSLGAGGVGVPVRATISSFG
Mflv_2544|M.gilvum_PYR-GCK SRPGEPHDKVAQRAADTGDEWAPTAYPVEFLSLGSAGLGVPVRATITSFG
Mvan_4099|M.vanbaalenii_PYR-1 SKPGEAGDKVTQRAADTGDEWAATAYPVEFLSLGTSGIGVPVRATISSFG
MSMEG_4723|M.smegmatis_MC2_155 AKPGESNDKVTQRAADTGDDWTPTGFPVEFLSLGTEGIGVPVRATITSFG
TH_1382|M.thermoresistible__bu ARPGEPGEKITQRAADTGDDWTPTGYPVEFLGLGTSGIGVPVRATVSSFG
MAB_1534|M.abscessus_ATCC_1997 SQPGEAGDKTAARAKDTGDDWVPTGFPTEFVSLGTNGIGVPVRATIRSFG
MAV_1665|M.avium_104 SRPGQGDDRTAARAKDTGDDWQATAFPVEFLSLGTNGIGVPIRATVESFG
::.*: :: : ** :*** * .*.:*.**:.**: *:***:***: ***
Mb2538c|M.bovis_AF2122/97 PILLAKVLGLNATQESTLGLIFHWADQRGLPLLDLKDLRAVITHLTSDEG
Rv2510c|M.tuberculosis_H37Rv PILLAKVLGLNATQESTLGLIFHWADQRGLPLLDLKDLRAVITHLTSDEG
MMAR_3858|M.marinum_M PILLAKVLGLNATQESTLGLIFHWADQRELPLLDLKDLRAVITHLISDEG
Mflv_2544|M.gilvum_PYR-GCK PVLLSKVLGLNQTQESTLGLIFHWADQQGLALLDLKDLRSVIQYLTSDEG
Mvan_4099|M.vanbaalenii_PYR-1 PILLSKVLGLNQTQESTLGLIFHWADQQGLALLDLKDLRAVIQHLTSDEG
MSMEG_4723|M.smegmatis_MC2_155 PILLSKVLGLNQTQESTLGLIFHWADQKGLPLLDIKDLRSVIQYLTSDEG
TH_1382|M.thermoresistible__bu PILLSKVLGLNQTQESTLGLICYWADQQGLPLLDLKDLRSVIQFLVSDEG
MAB_1534|M.abscessus_ATCC_1997 PILLSKVLGLNATQESTLGLIFHWADQQQLPLQDLKDLRAVITYLTSDEG
MAV_1665|M.avium_104 PVLLSKVLGLNATQESTLGLIFHWAKERNWPLVTTDNLRGAISHLTSDEG
*:**:****** ********* :**.:: .* .:**..* .* ****
Mb2538c|M.bovis_AF2122/97 KVELKSLGAVSPTTAGVILRALVNLEAEGADTFFGEPELRPEDLLRVDSQ
Rv2510c|M.tuberculosis_H37Rv KVELKSLGAVSPTTAGVILRALVNLEAEGADTFFGEPELRPEDLLRVDSQ
MMAR_3858|M.marinum_M KAELKSLGAVSPTTAGVILRALVNLEAEGADSFFGEPELRPDDLLRVDGQ
Mflv_2544|M.gilvum_PYR-GCK KPQLKALGAVSSTTAGVILRALVNLEAEGADTFFGEPELEPEDLMRVDAS
Mvan_4099|M.vanbaalenii_PYR-1 KPQLKALGAVSTTTAGVILRALVNLEAEGADTFFGEPELEPDDLLRVDAS
MSMEG_4723|M.smegmatis_MC2_155 KPELKALGAVSTTTAGVILRALVNLEAEGADTFFGEPELEPKDLLRVDDQ
TH_1382|M.thermoresistible__bu KPQLKSLGAVSPTTAGVILRALINLEADGGDTFFGEPELEPEDLLRVDDQ
MAB_1534|M.abscessus_ATCC_1997 KADLKNLGGVSAATAGVILRALINLEADDGDTFFGEPEIDPNDLMRTSGD
MAV_1665|M.avium_104 KEDLKSLGGVSATTAGVILRALVNLDAEGGDTFFGEPKLDPNDLLRTNDQ
* :** **.**.:*********:**:*:..*:*****:: *.**:*.. .
Mb2538c|M.bovis_AF2122/97 GRGIISLLEFGSQALRPAMFSTFLMWVLADLFTFLPEVGDLDKPKLVFFF
Rv2510c|M.tuberculosis_H37Rv GRGIISLLEFGSQALRPAMFSTFLMWVLADLFTFLPEVGDLDKPKLVFFF
MMAR_3858|M.marinum_M GRGIISLLELGSQSLRPVLFSTFLMWVLADLFTYLPEVGDLDKPKLVFFF
Mflv_2544|M.gilvum_PYR-GCK GRGIITLLELGPQAARPVLFSTFLMWVLADLFTTLPEVGDLDRPKLVFVF
Mvan_4099|M.vanbaalenii_PYR-1 GRGIITLLELGSQAARPVMFSTFLMWVLADLFTTLPEVGDVDKPKLVFVF
MSMEG_4723|M.smegmatis_MC2_155 GRGVITLLELGAQAARPVMFSTFLMWVLADLFTTLPEVGDVDKPKLVFFF
TH_1382|M.thermoresistible__bu GRGVITLFELGNQAARPVMFSTFLMWVLADLFTNLPEVGDLDKPKLVFFF
MAB_1534|M.abscessus_ATCC_1997 -KGVITLVELGAQAARPVLFSTFLMWILANLFTKLPEIGDVDKPKLVFIF
MAV_1665|M.avium_104 GQGVISLLEFTGQSVRPVIFSTFLMWLLADLYTQLPEVGDIDKPKLVFFF
:*:*:*.*: *: **.:*******:**:*:* ***:**:*:*****.*
Mb2538c|M.bovis_AF2122/97 DEAHLLFTDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDLPNDVLSQL
Rv2510c|M.tuberculosis_H37Rv DEAHLLFTDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDLPNDVLSQL
MMAR_3858|M.marinum_M DEAHLLFSDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDLPKDVLSQL
Mflv_2544|M.gilvum_PYR-GCK DEAHLLFADASKAFLDQVEQTVKLIRSKGVGVLFCTQLPTDIPKEVLGQL
Mvan_4099|M.vanbaalenii_PYR-1 DEAHLLFADASKAFLEQVEQTVKLIRSKGVGVLFCTQLPTDVPKEVLSQL
MSMEG_4723|M.smegmatis_MC2_155 DEAHLLFNDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDVPNDVLSQL
TH_1382|M.thermoresistible__bu DEAHLLFNDASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDVPNDVLSQL
MAB_1534|M.abscessus_ATCC_1997 DEAHLLFADASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDVPNNVLSQL
MAV_1665|M.avium_104 DEAHLLFADASKAFLEQVEQTVKLIRSKGVGVFFCTQLPTDIPNDVLSQL
******* *******:****************:********:*::**.**
Mb2538c|M.bovis_AF2122/97 GARIQHALRAFTPDDHKALRKTVRTYPKTDVYDLESALTSLGTGEAVVTV
Rv2510c|M.tuberculosis_H37Rv GARIQHALRAFTPDDHKALRKTVRTYPKTDVYDLESALTSLGTGEAVVTV
MMAR_3858|M.marinum_M GARIQHALRAFTPDDHKALTKTVRTYPKTDVYDLESALTSLGIGEAVVTV
Mflv_2544|M.gilvum_PYR-GCK GARIQHALRAFTPDDQKALTKTVRTYPKTTVYDLESALMSLGTGEAIVTV
Mvan_4099|M.vanbaalenii_PYR-1 GARIQHALRAFTPDDQKALTKTVRTYPKTAVYDLESALTSLGIGEAVVTV
MSMEG_4723|M.smegmatis_MC2_155 GARIQHALRAFTPDDQKALTKTVRTYPKTKVYDLEEALTSLGIGEAVVTV
TH_1382|M.thermoresistible__bu GARVQHALRAFTPDDQKALRKTVRTYPKTDVYDLESTLTSLGIGEAVVTV
MAB_1534|M.abscessus_ATCC_1997 GARVQHALRAFTPEDQKALAKTVKTYPKTDVYDMDTALTSLGIGEAIVTV
MAV_1665|M.avium_104 GARIQHALRAFTPDDQKALSKTVRTYPKTDVYDLESALTSLGIGEAVVTV
***:*********:*:*** ***:***** ***:: :* *** ***:***
Mb2538c|M.bovis_AF2122/97 LSEKGAPTPVAWTRMRAPRSLMAAIGAEAIGAAAQASSLQAVYGQTIDRP
Rv2510c|M.tuberculosis_H37Rv LSEKGAPTPVAWTRMRAPRSLMAAIGAEAIGAAAQASSLQAVYGQTIDRP
MMAR_3858|M.marinum_M LSEKGVPTPVAWTRMRAPRSLMGAIGSEAIRAVAQASPLQAVYGQTIDRE
Mflv_2544|M.gilvum_PYR-GCK LSERGAPTPVAWTRMRAPRSLMDTIGPDAITAAAHASPLQAEYGRAVDRE
Mvan_4099|M.vanbaalenii_PYR-1 LSERGAPTPVAWTRMRAPRSLMDTIGPEAIAAAAKASPLQAEYGQTIDRD
MSMEG_4723|M.smegmatis_MC2_155 LSEKGAPTPVAWTRMRAPRSLMDTIGADAISAAAKASPLQATYGQTVDRD
TH_1382|M.thermoresistible__bu LSEKGAPTPVARTLMRAPRSLMDTIGEDAIAAAAKSSPLQAEYGQTIDRE
MAB_1534|M.abscessus_ATCC_1997 LSERGAPTPVAWTRMRAPRSLMAAIGDDAIKAAAASSPLQSKYGQTVDSR
MAV_1665|M.avium_104 LSEKGAPTPVAWTRMRVPRSLMASIGTEEITKAAKRSELQAKYGQTVQQP
***:*.***** * **.***** :** : * .* * **: **::::
Mb2538c|M.bovis_AF2122/97 SAHEILSAKLAPAQEAPAQEAPAP---RGQYDPLPWPDDFEVPPMPAPVE
Rv2510c|M.tuberculosis_H37Rv SAHEILSAKLAPAQEAPAQEAPAP---RGQYDPLPWPDDFEVPPMPAPVE
MMAR_3858|M.marinum_M SAHEMLSAKYAPATEAAEPPATAPQPPRGKYDPLPWPQDFDLPPMPTPVE
Mflv_2544|M.gilvum_PYR-GCK SAYERLAARLAPPPPVVAEQ-----------PPVYVPPPGDLPPMPPMPA
Mvan_4099|M.vanbaalenii_PYR-1 SAHERLAAKLAPPPPVTPAPPASP-------PPVYIPPPVEVPPMPAPAQ
MSMEG_4723|M.smegmatis_MC2_155 SAYERLAAKLAPPEGGDAGG------------FAGVEATGEAAPMPAPAQ
TH_1382|M.thermoresistible__bu SAYERLAAALAPPEPEPTAPVPAP-------DLPPVPAGLDLPPMPAPVE
MAB_1534|M.abscessus_ATCC_1997 SAFEVLAEKAAQAQQD-------------------APVGQAPAKKNGKPA
MAV_1665|M.avium_104 AQPPPGPQAPTGSRSPQAQS-----------DYPPVPPNGPVPPMPEPAE
: . : . .
Mb2538c|M.bovis_AF2122/97 PQGPAVWEEILKNPTVKSVLN----TTAREITRSIFGTGRRRRK--
Rv2510c|M.tuberculosis_H37Rv PQGPAVWEEILKNPTVKSVLN----TTAREITRSIFGTGRRRRK--
MMAR_3858|M.marinum_M PQGPPLWEEVLKNPTVKSAIN----TAAREFTRSIFGTGRRRRK--
Mflv_2544|M.gilvum_PYR-GCK PAEPGLLDQMMANPAFKSAMRSAGTVIGREITRSIFGTGRRRRRR-
Mvan_4099|M.vanbaalenii_PYR-1 PDEPGLLDKMMDSPAFKSAMRSAGTVIGREITRSIFGTGRRRRRR-
MSMEG_4723|M.smegmatis_MC2_155 PEEPGFFEKMVASPAFKSAMRSAGTVIGREITRSIFGTGRRRR---
TH_1382|M.thermoresistible__bu PQGPGLLDKVMDSPAFKSAMRSAGTVIGREITRSIFGTGRRRRRRR
MAB_1534|M.abscessus_ATCC_1997 PEKPSWWRRLLSNRSFQSFLS----ALGKQLIR---NMGKK-----
MAV_1665|M.avium_104 PKGPGLLERLWHDRDVQSAVN----TGIREIIRLKFGGGGRRRK--
* * .: . .:* : . ::: * . * :