For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FETPLTVVGHIVNDPQRRKVGDQEFIKFRVASNSRRRTADGGWEPGNSLFITVNCWGRLVTGVGAALGKG APVIVVGHVYTSEYEDRDGNRRSSLELRATSVGPDLSRVIVRIEKPGYTGPSPEKAAAAAATTRCEDVAD NDDHSARDVADTADVAAGADAADRNALPLSA*
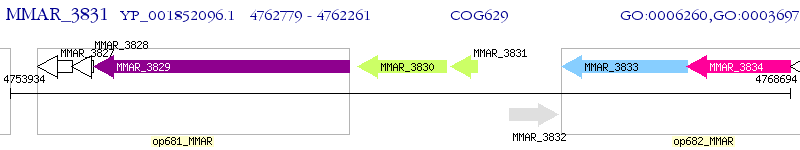
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3831 | - | - | 100% (172) | single-stranded DNA-binding protein |
| M. marinum M | MMAR_0073 | ssb | 7e-13 | 31.40% (121) | single-strand binding protein Ssb |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2505c | - | 1e-62 | 73.94% (165) | hypothetical protein Mb2505c |
| M. gilvum PYR-GCK | Mflv_2567 | - | 7e-54 | 62.79% (172) | hypothetical protein Mflv_2567 |
| M. tuberculosis H37Rv | Rv2478c | - | 1e-62 | 73.94% (165) | hypothetical protein Rv2478c |
| M. leprae Br4923 | MLBr_02684 | ssb | 3e-14 | 30.58% (121) | single-stranded DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_1559 | - | 5e-48 | 70.00% (130) | hypothetical protein MAB_1559 |
| M. avium 104 | MAV_1694 | ssb | 2e-67 | 75.58% (172) | hypothetical protein MAV_1694 |
| M. smegmatis MC2 155 | MSMEG_4701 | - | 5e-49 | 58.05% (174) | hypothetical protein MSMEG_4701 |
| M. thermoresistible (build 8) | TH_1034 | - | 7e-50 | 74.19% (124) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3758 | - | 3e-91 | 98.79% (165) | hypothetical protein MUL_3758 |
| M. vanbaalenii PYR-1 | Mvan_4072 | - | 3e-56 | 66.47% (167) | hypothetical protein Mvan_4072 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3831|M.marinum_M --MFETPLTVVGHIVNDPQRRKVGD-QEFIKFRVASNSRRR-TADGGWEP
MUL_3758|M.ulcerans_Agy99 ---------MVGHIVNDPQRRKVGD-QEFIKFRVASNSRRR-TADGGWEP
Mb2505c|M.bovis_AF2122/97 ---------MVGHIVNDLQRRKVGD-QEVVKFRVASNSRRR-TSDGGWEP
Rv2478c|M.tuberculosis_H37Rv ---------MVGHIVNDLQRRKVGD-QEVVKFRVASNSRRR-TSDGGWEP
MAV_1694|M.avium_104 --MFETPLTVVGHIVNNPERRQVGA-QEVIKFRVASNSRRR-TADGGWEP
MAB_1559|M.abscessus_ATCC_1997 --MFETPVTVIGSIVGDLKRRQVGS-DEMIKFRVASNARRR-REDGSWVN
Mflv_2567|M.gilvum_PYR-GCK --MFETPITIVGNIITAPDRRWVGD-QELIKFRVASNSRRR-TAEGTWEP
Mvan_4072|M.vanbaalenii_PYR-1 --MFETPITVVGTIVTQPDRRWVGD-QELFKFRVASNSRRR-TADGTWEP
TH_1034|M.thermoresistible__bu --MFETPITVVGNIVTKPDRRWVGD-QELIKFRMASNSRRR-NAAGEWEP
MSMEG_4701|M.smegmatis_MC2_155 MNMFETPFTVVGNIITNPVRLRFGD-QELYKFRVASNSRRR-SPEGTWEP
MLBr_02684|M.leprae_Br4923 -MAGDTTITIVGNLTADPELRFTSSGAAVVNFTVASTPRIYDRQSGEWKD
::* : . . :* :**..* * *
MMAR_3831|M.marinum_M GNSLFITVNCWGRLVTGVGAALGKGAPVIVVGHVYTSEYEDRDGNRRSSL
MUL_3758|M.ulcerans_Agy99 GNSLFITVNCWGRLVTGVGAALGKGAPVIVVGHVYTSEYEDRDGNRRSSL
Mb2505c|M.bovis_AF2122/97 GNSLFITVNCWGRLVTGVGAALGKGAPVIVVGHVYTSEYEDRDGIRRSSL
Rv2478c|M.tuberculosis_H37Rv GNSLFITVNCWGRLVTGVGAALGKGAPVIVVGHVYTSEYEDRDGIRRSSL
MAV_1694|M.avium_104 GNSLFINVNCWGRLVTGVGAALGKGAPVIVVGHVYTSEYEDRDGNRRSSL
MAB_1559|M.abscessus_ATCC_1997 SNSLFINVTCWGRLVTGVGAALGKGAPVIVHGHLHTSEFEDKDGNRRQVT
Mflv_2567|M.gilvum_PYR-GCK GNSLYATVNCWGNLITGVGAGLGKGDPVIVVGNVYTNEYEDRDGNRRSSL
Mvan_4072|M.vanbaalenii_PYR-1 GNSLYATVNCWGRLVTGVGAGLGKGDPVIVVGYLYTSEYEDRDGNRRSSL
TH_1034|M.thermoresistible__bu GNTLFVTVNCWGRLVTGVGAALGKGDAVIVVGYVYTSEYEDREGVRRSSV
MSMEG_4701|M.smegmatis_MC2_155 GNSLYVTVNCWGNLARGVSASLGKGDSVVVVGHLYTNEYEDREGVRRSSV
MLBr_02684|M.leprae_Br4923 GEALFLRCNIWREAAENVAESLTRGARVIVTGRLKQRSFETREGEKRTVV
.::*: . * . .*. .* :* *:* * : .:* ::* :*
MMAR_3831|M.marinum_M ELRATSVGPDLSRVIVRIEKPGYTGPSPEKAAAAAATTRCEDVADNDDHS
MUL_3758|M.ulcerans_Agy99 ELRATSVGPDLSRVIVRIEKPGYTGPSPGKAAAAAATTRCEDVADNDDHS
Mb2505c|M.bovis_AF2122/97 EMRATSVGPDLSRVIVRIEKPAYTGPSAGDLPAATGTG----AAGAADAP
Rv2478c|M.tuberculosis_H37Rv EMRATSVGPDLSRVIVRIEKPAYTGPSAGDLPAATGTG----AAGAADAP
MAV_1694|M.avium_104 EMRATSVGPDVSRAIVRIEKPGYTGPSTDEAAPA--------AAEAADAA
MAB_1559|M.abscessus_ATCC_1997 EMKAIAVGPDLSRTIARIEKVGYTG-KPDDAEAA----------EQPDPG
Mflv_2567|M.gilvum_PYR-GCK EIRATSVGPDLARCIVRMESRRHSRPAVTEPRDEADGRDTADAEDAEDAV
Mvan_4072|M.vanbaalenii_PYR-1 EIRATSVGPDLARCIIRIESRRYPEPVPAAVAVASEADSVDDGDVVGDVV
TH_1034|M.thermoresistible__bu EVKATSVGPDLARCVARIEHRRHPEPGPAGDRQQSAGPHNAANGVGPDPG
MSMEG_4701|M.smegmatis_MC2_155 EVRATAVGPDLSRCIARVEKVQPSQGPRADAGGDPE------RVPDDDVD
MLBr_02684|M.leprae_Br4923 EVEVDEIGPSLRYATAKVNKASRSGGGGGGFGSGSRQ----APAQMSGGV
*:.. :**.: ::: . .
MMAR_3831|M.marinum_M ARDVADTADVAAGADAADRNA----LPLSA
MUL_3758|M.ulcerans_Agy99 ARDVADTADVAAGADAADGNA----LPLSA
Mb2505c|M.bovis_AF2122/97 ASAADSVSDVVVDDAITGHNP----LPISA
Rv2478c|M.tuberculosis_H37Rv ASAADSVSDVVVDDAITGHNP----LPISA
MAV_1694|M.avium_104 GEDADRGDTVEP----VDTGP----VPLSA
MAB_1559|M.abscessus_ATCC_1997 AEES-AVVQPEGGESSEESVN----LRLSA
Mflv_2567|M.gilvum_PYR-GCK ETPSAER-------DDE-------ALSLSA
Mvan_4072|M.vanbaalenii_PYR-1 DEDASGS-------GAE-------ELSLSA
TH_1034|M.thermoresistible__bu EEPPGEEPEVEPTEGAEPDPPEGAGLPLTA
MSMEG_4701|M.smegmatis_MC2_155 RRDADDEPDDLADDDVADLAG--DGLPLTA
MLBr_02684|M.leprae_Br4923 GDDPWGSAPTSGSFGVGDEEP-----PF--
: