For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
WRMRVVHSTGYAYEAPVTASFNEARLTPRSNSRQNVILNRVETIPATRSYRYVDYWGTAVTAFDLHAPHT ELTVTSSSVVETERPEPPEAKVTWEELESMAVIDRFDEVLRPTSYTPTSKRVAAVGRRIRKYHDPAEAVV AAARWARSELDYLPGTTGVHSSGLDALEQGKGVCQDFAHLSLIVLRGMGIPARYVSGYLHPKRNAVVGKS VAGRSHAWIQAWTGGWWHYDPTNDTEISEQYISVGAGRDYSDVPPLKGIYSGLGVTDLDVDVEITRLA*
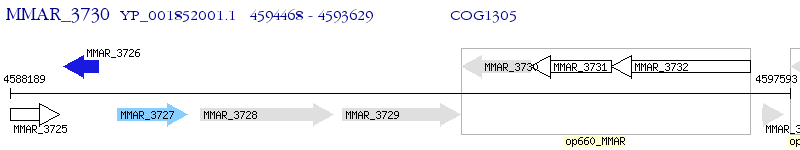
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3730 | - | - | 100% (279) | putative transglutaminase-like protein |
| M. marinum M | MMAR_2162 | - | 1e-32 | 33.56% (295) | hypothetical protein MMAR_2162 |
| M. marinum M | MMAR_2165 | - | 5e-08 | 24.09% (303) | transglutaminase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2432c | - | 1e-142 | 84.95% (279) | hypothetical protein Mb2432c |
| M. gilvum PYR-GCK | Mflv_2668 | - | 1e-134 | 79.93% (279) | transglutaminase domain-containing protein |
| M. tuberculosis H37Rv | Rv2409c | - | 1e-142 | 84.95% (279) | hypothetical protein Rv2409c |
| M. leprae Br4923 | MLBr_00607 | - | 1e-138 | 82.80% (279) | hypothetical protein MLBr_00607 |
| M. abscessus ATCC 19977 | MAB_1637 | - | 1e-125 | 72.76% (279) | transglutaminase-like |
| M. avium 104 | MAV_1773 | - | 1e-130 | 77.06% (279) | hypothetical protein MAV_1773 |
| M. smegmatis MC2 155 | MSMEG_4567 | - | 1e-134 | 80.29% (279) | hypothetical protein MSMEG_4567 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3673 | - | 1e-164 | 100.00% (279) | putative transglutaminase-like protein |
| M. vanbaalenii PYR-1 | Mvan_3909 | - | 1e-134 | 78.85% (279) | transglutaminase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2668|M.gilvum_PYR-GCK MWRMRVVHSTGYAYKSPVTASYNEARLTPRSDSRQNVILNRVETVPATRS
Mvan_3909|M.vanbaalenii_PYR-1 MWRMRVVHSTGYAYKSPVTASFNEARLTPRSDSRQNVILNRVETVPATRS
Mb2432c|M.bovis_AF2122/97 MWRTRVVHTTGYVYQSPVTASYNEARLTPRSSSRQNLVLNRVETIPATRS
Rv2409c|M.tuberculosis_H37Rv MWRTRVVHTTGYVYQSPVTASYNEARLTPRSSSRQNLVLNRVETIPATRS
MMAR_3730|M.marinum_M MWRMRVVHSTGYAYEAPVTASFNEARLTPRSNSRQNVILNRVETIPATRS
MUL_3673|M.ulcerans_Agy99 MWRMRVVHSTGYAYEAPVTASFNEARLTPRSNSRQNVILNRVETIPATRS
MLBr_00607|M.leprae_Br4923 MWRMRVVHSTGYVYESPATASFNEARLTPRSNTRQTVILNRVETIPATRS
MAV_1773|M.avium_104 MWRLRVVHTTGYAYQSPVTASYNEARLTPRSNNRQNVILNRVETIPATRS
MSMEG_4567|M.smegmatis_MC2_155 MWRLRVVHSTGYAYKSPVTASFNEVRLTPRSDSRQNVILNRVETVPATRS
MAB_1637|M.abscessus_ATCC_1997 MWRLRIMHATGYAYKTPVTASYNEARLTPRSDGRQNVILNRVETVPATRS
*** *::*:***.*::*.***:**.******. **.::******:*****
Mflv_2668|M.gilvum_PYR-GCK YRYVDYWGTAVTAFDLHAPHTELEVTASSVVETDKPEQPDVKVTWEDLSS
Mvan_3909|M.vanbaalenii_PYR-1 YRYVDYWGTAVTAFDLHAPHTELEVTASSVVETDRPEEPKTKATWADLAT
Mb2432c|M.bovis_AF2122/97 YRYIDYWGTAVTAFDLHAPHTELTVTSSSVVETERPEPLAAKATWADLQS
Rv2409c|M.tuberculosis_H37Rv YRYIDYWGTAVTAFDLHAPHTELTVTSSSVVETERPEPLAAKATWADLQS
MMAR_3730|M.marinum_M YRYVDYWGTAVTAFDLHAPHTELTVTSSSVVETERPEPPEAKVTWEELES
MUL_3673|M.ulcerans_Agy99 YRYVDYWGTAVTAFDLHAPHTELTVTSSSVVETERPEPPEAKVTWEELES
MLBr_00607|M.leprae_Br4923 YRYIDYWGTVVTAFDLRAPHTELTVTSSSVVETESLEPSVSRITWTELQS
MAV_1773|M.avium_104 YRYIDYWGTAVTAFDLHAPHTDLTVMSSSVVETERAERPATELGWGDLHS
MSMEG_4567|M.smegmatis_MC2_155 YRYVDYWGTAVTAFDLHAPHAELEVTSSSVVETEVGEKPEEPVSWDDLRT
MAB_1637|M.abscessus_ATCC_1997 YRYVDYWGTAVTTFDLHAPHTELEVTGLSVVETDVAEKPEEVVDWDEVHT
***:*****.**:***:***::* * . *****: * * :: :
Mflv_2668|M.gilvum_PYR-GCK EAVIDRYDEVLAPTHYVPASKRIERVGRRIAKYHEPGEAVIEAARWVQGE
Mvan_3909|M.vanbaalenii_PYR-1 EAVVDKYDEVLAPTHYAPSSKRIERVGRRIAKYHEPAEAVLEAARWVQGE
Mb2432c|M.bovis_AF2122/97 TAVIDRFDEVLRPTPHTPASARVDAVGRRIRKCHEPSEAVVAAARWARSE
Rv2409c|M.tuberculosis_H37Rv TAVIDRFDEVLRPTPHTPASARVDAVGRRIRKCHEPSEAVVAAARWARSE
MMAR_3730|M.marinum_M MAVIDRFDEVLRPTSYTPTSKRVAAVGRRIRKYHDPAEAVVAAARWARSE
MUL_3673|M.ulcerans_Agy99 MAVIDRFDEVLRPTSYTPTSKRVAAVGRRIRKYHDPAEAVVAAARWARSE
MLBr_00607|M.leprae_Br4923 MAVIDRFDEVLQPTAYTPVNTRVAAVGKRIAKNHEPRKAVVAAARWARSK
MAV_1773|M.avium_104 EAVIDRFDELLRPTGHVPASKRVASVAKKIAKSHDPAEAVVAVGDWVRGE
MSMEG_4567|M.smegmatis_MC2_155 EAVIDRFDEVLAPTHYTPCSKRIERVGRSIAKEHDPYEAVIAAAQWVRSE
MAB_1637|M.abscessus_ATCC_1997 TAVVDRFDEYLSPTAYVPQSKKVQSVGKRIAKDVSPAEAIIAASKWVHSE
**:*::** * ** :.* . :: *.: * * .* :*:: .. *.:.:
Mflv_2668|M.gilvum_PYR-GCK LQYVPGTTGVHTSGVDALREGKGVCQDFAHLTLMVLRGMGIPSRYVSGYL
Mvan_3909|M.vanbaalenii_PYR-1 LQYVPGTTGVHTSAVDALREGKGVCQDFAHLTLMLLRGMGIPSRYVSGYL
Mb2432c|M.bovis_AF2122/97 LDYIPGTTSVHSSGLDALEQGKGVCQDFVHLSLMVLRSMGIPCRYVSGYL
Rv2409c|M.tuberculosis_H37Rv LDYIPGTTSVHSSGLDALEQGKGVCQDFVHLSLMVLRSMGIPCRYVSGYL
MMAR_3730|M.marinum_M LDYLPGTTGVHSSGLDALEQGKGVCQDFAHLSLIVLRGMGIPARYVSGYL
MUL_3673|M.ulcerans_Agy99 LDYLPGTTGVHSSGLDALEQGKGVCQDFAHLSLIVLRGMGIPARYVSGYL
MLBr_00607|M.leprae_Br4923 LDYIPGTTGVHSSGLDALHQCRGVCQDFAHLTLIVLRSMGIPGRYVSGYL
MAV_1773|M.avium_104 LEYLPGTTSVHSSGLDALKEGRGVCQDFVHLSLMLLRSMGIPSHYVSGYL
MSMEG_4567|M.smegmatis_MC2_155 LDYVPGTTGVHSSGLDALREGKGVCQDFAHLSLIMLRGMGIPARYVSGYL
MAB_1637|M.abscessus_ATCC_1997 LDYMPGTTGVHSSALDALRESKGVCQDYAHLTLVLLRSMGVPARYVSGYL
*:*:****.**:*.:***.: :*****:.**:*::**.**:* :******
Mflv_2668|M.gilvum_PYR-GCK HPKKNAKVGDTVDGQSHAWIQAWTGEWRYYDPTNDAEINEQYISVGVGRD
Mvan_3909|M.vanbaalenii_PYR-1 HPKRNAKIGDTVEGQSHAWIQAWTGEWWHYDPTNDADINEQYISVGVGRD
Mb2432c|M.bovis_AF2122/97 HPKRDAVVGKTVDGRSHAWVQAWTGGWWHYDPTNDNEITEQYISVGVGRD
Rv2409c|M.tuberculosis_H37Rv HPKRDAVVGKTVDGRSHAWVQAWTGGWWHYDPTNDNEITEQYISVGVGRD
MMAR_3730|M.marinum_M HPKRNAVVGKSVAGRSHAWIQAWTGGWWHYDPTNDTEISEQYISVGAGRD
MUL_3673|M.ulcerans_Agy99 HPKRNAVVGKSVAGRSHAWIQAWTGGWWHYDPTNDTEISEQYISVGAGRD
MLBr_00607|M.leprae_Br4923 HPKRDAVVGKTVEGRSHAWIQAWTGGWWNYDPTNDVEITEQYISVGVGRD
MAV_1773|M.avium_104 HPKRDAVVGDTVDGRSHAWIEAWTGAWWSYDPTNDAEITEQYVGVGAGRD
MSMEG_4567|M.smegmatis_MC2_155 HPTRGAAVGDTIDGQSHAWVQAWTGGWWDYDPTNDSEINEQYVSVGVGRN
MAB_1637|M.abscessus_ATCC_1997 HPKRDAAVGETVDGQSHAWIQGWAGGWWDYDPTNDSEINEQYVTVGVGRD
**.:.* :*.:: *:****::.*:* * ****** :*.***: **.**:
Mflv_2668|M.gilvum_PYR-GCK YSDVTPLKGIYSGEGSTDLDVIVEITRLA---------------------
Mvan_3909|M.vanbaalenii_PYR-1 YSDVTPLKGIYSGEGSTDLDVIVEITRLA---------------------
Mb2432c|M.bovis_AF2122/97 YTDVSPLKGIYSGEGVTDLDVVVEITRLA---------------------
Rv2409c|M.tuberculosis_H37Rv YTDVSPLKGIYSGEGVTDLDVVVEITRLA---------------------
MMAR_3730|M.marinum_M YSDVPPLKGIYSGLGVTDLDVDVEITRLA---------------------
MUL_3673|M.ulcerans_Agy99 YSDVPPLKGIYSGLGVTDLDVDVEITRLA---------------------
MLBr_00607|M.leprae_Br4923 YTDVAPLKGIYSGRGATDLDVVVEITRLA---------------------
MAV_1773|M.avium_104 YADVSPLKGIYSGKGATDLDVIVEVTRLALPTEEPAAIRRNAWTTPRAAP
MSMEG_4567|M.smegmatis_MC2_155 YSDVAPLKGIYSGEGSTDLDVVVEITRLA---------------------
MAB_1637|M.abscessus_ATCC_1997 YSDVSPLKGIYSGGGSTDLDVVVEVTRLA---------------------
*:**.******** * ***** **:****
Mflv_2668|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3909|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2432c|M.bovis_AF2122/97 --------------------------------------------------
Rv2409c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3730|M.marinum_M --------------------------------------------------
MUL_3673|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00607|M.leprae_Br4923 --------------------------------------------------
MAV_1773|M.avium_104 PGCRCCRCSASACWAAGSPSGSGWAASCRCRTARWPRWRPMFAPSRMPCG
MSMEG_4567|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1637|M.abscessus_ATCC_1997 --------------------------------------------------