For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPRETRAADAVGARPADPQVRSSIDVPPDLVVGLLGSADENLRALERTLTASLHVRGNTVTLAGEPADVA IAERAVSELVAIVANGQPLTPEVVRHSVAMLVGAGNESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAID ANTIVFGIGPAGTGKTYLAMAKAVHALQTKQVNRIILTRPAVEAGERLGFLPGTLSEKIDPYLRPLYDAL YDMMDPELIAKLMSAGVIEVAPLAYMRGRTLNDAFIVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQ VDLPGGARSGLRAAVDILDDIDDIHIAELTSADVVRHRLVSEIVDAYARYEEPGSGMNRAARRASGTRNR R*
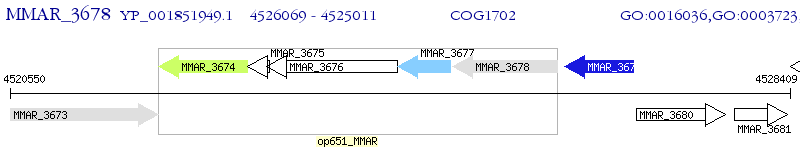
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3678 | phoH1 | - | 100% (352) | PhoH-like protein PhoH1 |
| M. marinum M | MMAR_4372 | phoH2 | 6e-26 | 30.74% (270) | PhoH-like protein PhoH2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2389c | phoH1 | 0.0 | 93.47% (352) | phosphate starvation-inducible protein PsiH |
| M. gilvum PYR-GCK | Mflv_2706 | - | 1e-152 | 79.49% (351) | PhoH family protein |
| M. tuberculosis H37Rv | Rv2368c | phoH1 | 0.0 | 93.47% (352) | phosphate starvation-inducible protein PsiH |
| M. leprae Br4923 | MLBr_00627 | phoH | 1e-169 | 87.50% (352) | phoH-family protein |
| M. abscessus ATCC 19977 | MAB_1668 | - | 1e-152 | 81.27% (331) | PhoH-like protein |
| M. avium 104 | MAV_2025 | - | 1e-172 | 86.65% (352) | PhoH family protein |
| M. smegmatis MC2 155 | MSMEG_4497 | - | 1e-160 | 82.39% (352) | PhoH family protein |
| M. thermoresistible (build 8) | TH_1054 | phoH | 1e-158 | 82.10% (352) | PROBABLE PHOH-LIKE PROTEIN PHOH1 (PHOSPHATE |
| M. ulcerans Agy99 | MUL_3621 | phoH1 | 0.0 | 99.72% (352) | PhoH-like protein PhoH1 |
| M. vanbaalenii PYR-1 | Mvan_3832 | - | 1e-154 | 79.55% (352) | PhoH family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2706|M.gilvum_PYR-GCK MAPRE-NASSDASQSSAR--SSSRIDVPPGLIMGLLGSSDENLRALERLL
Mvan_3832|M.vanbaalenii_PYR-1 MAPRE-NAGSDASAASSEPLVRSTITVPPEYVMGLLGSSDENLRELEQLL
MSMEG_4497|M.smegmatis_MC2_155 MTPR--DTNADSTATAQES-VRSSITVPPDIIVGLLGSADENLRAIEKVL
TH_1054|M.thermoresistible__bu VTPRETSAGSDASVAAVR----SSITVPPDLVVGLLGSADENLRTIEDLL
MMAR_3678|M.marinum_M MTPRETRAADAVGARPADPQVRSSIDVPPDLVVGLLGSADENLRALERTL
MUL_3621|M.ulcerans_Agy99 MTPRETRAADAVGARPADPQVRSSIDVPPDLVVGLLGSADENLRALERTL
Mb2389c|M.bovis_AF2122/97 MTSRETRAADAAGARQADAQVRSSIDVPPDLVVGLLGSADENLRALERTL
Rv2368c|M.tuberculosis_H37Rv MTSRETRAADAAGARQADAQVRSSIDVPPDLVVGLLGSADENLRALERTL
MLBr_00627|M.leprae_Br4923 MTHHETSAVDAS---SASMHVRSTIDVPPDLVVGLLGSADENLRALERML
MAV_2025|M.avium_104 MTPRDTSAADAAGALQADAQVRISIDVPPDIVMGLLGSADENLRALERSV
MAB_1668|M.abscessus_ATCC_1997 MPSVST------------ADVRSSVEIPNDLIMGLLGSADENLRELEDVL
:. : :* ::*****:***** :* :
Mflv_2706|M.gilvum_PYR-GCK TADIHVRGNAISLSGEPDDVALAERAVSELVAVVASGQTLTPDAVRHTVA
Mvan_3832|M.vanbaalenii_PYR-1 AADIHARGNEITVAGDHADVALADRAISELIEVVGSGQRLTPDAVRHSVA
MSMEG_4497|M.smegmatis_MC2_155 TSDVHVRGNEVTFSGEPADVALAERVVAELVAVASSGQAVTPETVRHSVA
TH_1054|M.thermoresistible__bu AADLHVRGNAITFTGESADVALAERVISELVTIVGSGHPLTPDAIRHSVS
MMAR_3678|M.marinum_M TASLHVRGNTVTLAGEPADVAIAERAVSELVAIVANGQPLTPEVVRHSVA
MUL_3621|M.ulcerans_Agy99 TASLHVRGNTVTLAGEPADVAIAERAVSELVAIVANGQPLTPEVVRHSVA
Mb2389c|M.bovis_AF2122/97 SADLHVRGNAVTLCGEPADVALAERVISELIAIVASGQSLTPEVVRHSVA
Rv2368c|M.tuberculosis_H37Rv SADLHVRGNAVTLCGEPADVALAERVISELIAIVASGQSLTPEVVRHSVA
MLBr_00627|M.leprae_Br4923 SADLHVRGNTVAFSGEPADVALAERAISELVAIVASGHPLTPEVVRHGVA
MAV_2025|M.avium_104 IADLHVRGNAITISGESADVARAERVISELVAIVANGQVLTPEVVRHSVA
MAB_1668|M.abscessus_ATCC_1997 PADVHARGNTITFTGVPAEVALAERVISELIVIARRGQQLTPSTVRHTVA
:.:*.*** ::. * :** *:*.::**: :. *: :**..:** *:
Mflv_2706|M.gilvum_PYR-GCK MLAGMQDGTVEESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDTHTIV
Mvan_3832|M.vanbaalenii_PYR-1 MLTGADD----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDAKTIV
MSMEG_4497|M.smegmatis_MC2_155 ILTGAGD----ESPAEVLTLDILSRRGKTIRPKTLNQKHYVDAIDAHTIV
TH_1054|M.thermoresistible__bu MLTGSGN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDAHTIV
MMAR_3678|M.marinum_M MLVGAGN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
MUL_3621|M.ulcerans_Agy99 MLVGAGN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
Mb2389c|M.bovis_AF2122/97 MLVGTGN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
Rv2368c|M.tuberculosis_H37Rv MLVGTGN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
MLBr_00627|M.leprae_Br4923 MLVGTGN----ESPAEVLTLDILLRRGKTVRPKTLNQKRYVDAIDANTIV
MAV_2025|M.avium_104 MLAGTDN----ESPAEVLTLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
MAB_1668|M.abscessus_ATCC_1997 MLTGDGD----ESPAEVLSLDILSRRGKTIRPKTLNQKRYVDAIDANTIV
:*.* : *******:**** *****:********:******::***
Mflv_2706|M.gilvum_PYR-GCK FGIGPAGTGKTYLAMAKAVSALQGKQVSRIILTRPAVEAGERLGFLPGTL
Mvan_3832|M.vanbaalenii_PYR-1 FGIGPAGTGKTYLAMAKAVSALQAKQVSRIILTRPAVEAGERLGFLPGTL
MSMEG_4497|M.smegmatis_MC2_155 FGIGPAGTGKTYLAMAKAVNALQTKQVNRIILTRPAVEAGERLGFLPGTL
TH_1054|M.thermoresistible__bu FGVGPAGTGKTYLAMAKAVSALQAKQVNRIILTRPAVEAGERLGFLPGTL
MMAR_3678|M.marinum_M FGIGPAGTGKTYLAMAKAVHALQTKQVNRIILTRPAVEAGERLGFLPGTL
MUL_3621|M.ulcerans_Agy99 FGIGPAGTGKTYLAMAKAVHALQTKQVNRIILTRPAVEAGERLGFLPGTL
Mb2389c|M.bovis_AF2122/97 FGIGPAGTGKTYLAMAKAVHALQTKQVTRIILTRPAVEAGERLGFLPGTL
Rv2368c|M.tuberculosis_H37Rv FGIGPAGTGKTYLAMAKAVHALQTKQVTRIILTRPAVEAGERLGFLPGTL
MLBr_00627|M.leprae_Br4923 FGIGPAGTGKTYLAMAKAVNALQTKQVTRIILTRPAVEAGERLGFLPGTL
MAV_2025|M.avium_104 FGVGPAGTGKTYLAMAKAVNALQTKQVSRIILTRPAVEAGERLGFLPGTL
MAB_1668|M.abscessus_ATCC_1997 FGIGPAGTGKTYLAMAKAVNALQSKQVTRIILTRPAVEAGERLGFLPGTL
**:**************** *** ***.**********************
Mflv_2706|M.gilvum_PYR-GCK SEKIDPYLRPLYDALHDMMDPELIPKLMSAGVIEVAPLAYMRGRSLNDAF
Mvan_3832|M.vanbaalenii_PYR-1 YEKIDPYLRPLYDALHDMMDPELIPKLMTAGVIEVAPLAFMRGRTLNDAF
MSMEG_4497|M.smegmatis_MC2_155 SEKIDPYLRPLYDALHDMMDTELIPKLMSAGVIEVAPLAYMRGRTLNDAF
TH_1054|M.thermoresistible__bu SEKIDPYLRPLYDALHDMMDPELIPKLMSAGVIEVAPLGYMRGRTLNDAF
MMAR_3678|M.marinum_M SEKIDPYLRPLYDALYDMMDPELIAKLMSAGVIEVAPLAYMRGRTLNDAF
MUL_3621|M.ulcerans_Agy99 SEKIDPYLRPLYDALYDMMDPELIAKLMSAGVIEVAPLAYMRGRTLNDAF
Mb2389c|M.bovis_AF2122/97 SEKIDPYLRPLYDALYDMMDPELIPKLMSAGVIEVAPLAYMRGRTLNDAF
Rv2368c|M.tuberculosis_H37Rv SEKIDPYLRPLYDALYDMMDPELIPKLMSAGVIEVAPLAYMRGRTLNDAF
MLBr_00627|M.leprae_Br4923 SEKIDPYLRPLYDALYDMMDPELIPKLMSSGVIEVASLAYMRGRTLNDAF
MAV_2025|M.avium_104 SEKIDPYLRPLYDALYDMMDPEVIPKLMSAGVIEVAPLAYMRGRTLNSAF
MAB_1668|M.abscessus_ATCC_1997 SEKIDPYLRPLYDALHDMMDPELIPKLMDAGVIEVAPLAYMRGRTINDAF
**************:****.*:*.*** :******.*.:****::*.**
Mflv_2706|M.gilvum_PYR-GCK IILDEAQNTTGEQMKMFLTRLGFGSKMVVTGDITQVDLPGGAPSGLNTAM
Mvan_3832|M.vanbaalenii_PYR-1 IILDEAQNTTAEQMKMFLTRLGFGSKIVVTGDITQVDLPGGAASGLRAAM
MSMEG_4497|M.smegmatis_MC2_155 IILDEAQNTTAEQMKMFLTRLGFGSKIVVTGDVTQVDLPGGATSGLRAAM
TH_1054|M.thermoresistible__bu IILDEAQNTTAEQMKMFLTRLGFGSKIVVTGDVTQVDLPGGATSGLRAAM
MMAR_3678|M.marinum_M IVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQVDLPGGARSGLRAAV
MUL_3621|M.ulcerans_Agy99 IVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQVDLPGGARSGLRAAV
Mb2389c|M.bovis_AF2122/97 IVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQIDLPGGARSGLRAAV
Rv2368c|M.tuberculosis_H37Rv IVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQIDLPGGARSGLRAAV
MLBr_00627|M.leprae_Br4923 IVLDEAQNTTAEQMKMFLTRLGFGSKVVVTGDVTQIDLPGGARSGLRAAV
MAV_2025|M.avium_104 IVLDEAQNTTAEQMKMFLTRLGFGSKIVVTGDITQVDLPGGATSGLRSAM
MAB_1668|M.abscessus_ATCC_1997 IILDEAQNTTAEQMKMFLTRLGFGSKIVVTGDITQVDLPGNARSGLRAAM
*:********.***************:*****:**:****.* ***.:*:
Mflv_2706|M.gilvum_PYR-GCK RVLGDVDDIHFAELTSADVVRHRLVSEIVDAYSKFEEPTL-MNRAQRRAS
Mvan_3832|M.vanbaalenii_PYR-1 RILDGIDDIHFAELTSADVVRHRLVSEIVDAYAKAEEPTL-MNRAQRRSS
MSMEG_4497|M.smegmatis_MC2_155 QILDNIDDIHFAELTSADVVRHRLVSDIVDAYARHEEPAM-LNRAQRRSS
TH_1054|M.thermoresistible__bu KILGDIDDIHFSELTSADVVRHRLVSEIVDAYEKAETPAL-MNRAQRRAA
MMAR_3678|M.marinum_M DILDDIDDIHIAELTSADVVRHRLVSEIVDAYARYEEPGSGMNRAARRAS
MUL_3621|M.ulcerans_Agy99 DILDDIDDIHIAELNSADVVRHRLVSEIVDAYARYEEPGSGMNRAARRAS
Mb2389c|M.bovis_AF2122/97 DILEDIDDIHIAELTSVDVVRHRLVSEIVDAYARYEEPGSGLNRAARRAS
Rv2368c|M.tuberculosis_H37Rv DILEDIDDIHIAELTSVDVVRHRLVSEIVDAYARYEEPGSGLNRAARRAS
MLBr_00627|M.leprae_Br4923 DILEHIDDIYVAELASVDVVRHRLVSEIVDAYAEYEEHDLGMNRAARRAS
MAV_2025|M.avium_104 EILDSVDDIHVAELTSVDVVRHRLVSEIVDAYAKFEEPGLTMNRAARRAS
MAB_1668|M.abscessus_ATCC_1997 DILGNIEGIHFSELTSADVVRHRLVSEIVDAYARFEDSDLTGNRAQRRAS
:* ::.*:.:** *.*********:***** . * *** **::
Mflv_2706|M.gilvum_PYR-GCK GPGRPRR-
Mvan_3832|M.vanbaalenii_PYR-1 GSGRPRR-
MSMEG_4497|M.smegmatis_MC2_155 N-GRPRR-
TH_1054|M.thermoresistible__bu G-SRPRR-
MMAR_3678|M.marinum_M G-TRNRR-
MUL_3621|M.ulcerans_Agy99 G-TRNRR-
Mb2389c|M.bovis_AF2122/97 G-ARGRR-
Rv2368c|M.tuberculosis_H37Rv G-ARGRR-
MLBr_00627|M.leprae_Br4923 G-ARNRR-
MAV_2025|M.avium_104 G-SRGRR-
MAB_1668|M.abscessus_ATCC_1997 G-NRSRHR
. * *: