For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HRHLRVILMILGLVLGLGAPGCGHKSAPRPTTAPDTCKNSDGPTSQTVRQAIAAVPVGVPNSTWVEIARG HTKNCRLYWVQIIPTIASESTPQQLLFFDHNTALGSPTRNPKPYIRVLPSGQDTVTVQYQWRVGSDPQCC PTGSGTVRFQVGPDGKLEALGPIPHQ*
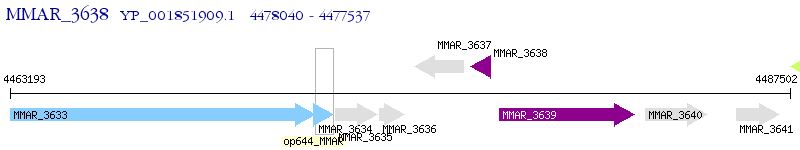
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3638 | lppP | - | 100% (167) | lipoprotein LppP |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2357c | lppP | 5e-69 | 71.78% (163) | lipoprotein LppP |
| M. gilvum PYR-GCK | Mflv_2725 | - | 2e-29 | 43.71% (151) | LppP |
| M. tuberculosis H37Rv | Rv2330c | lppP | 5e-69 | 71.78% (163) | lipoprotein LppP |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_2076 | - | 4e-62 | 68.83% (154) | LppP protein |
| M. smegmatis MC2 155 | MSMEG_4475 | - | 4e-38 | 46.50% (157) | LppP protein |
| M. thermoresistible (build 8) | TH_0726 | lppP | 1e-45 | 53.46% (159) | PROBABLE LIPOPROTEIN LPPP |
| M. ulcerans Agy99 | MUL_1475 | lppP | 1e-97 | 98.20% (167) | lipoprotein LppP |
| M. vanbaalenii PYR-1 | Mvan_3809 | - | 2e-27 | 40.76% (157) | LppP |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3638|M.marinum_M MHR--------HLRVILMILGLVLGLGAPGCGHKSAPRPTTAPDTCKNSD
MUL_1475|M.ulcerans_Agy99 MHR--------HLRVILMILGLVLGLGAPGCGHKSAPRPTTAPDTCKNSD
Mb2357c|M.bovis_AF2122/97 MRRQRSAVPILALLALLALLALIVGLGASGCAWKPPTTRPSPPNTCKDSD
Rv2330c|M.tuberculosis_H37Rv MRRQRSAVPILALLALLALLALIVGLGASGCAWKPPTTRPSPPNTCKDSD
MAV_2076|M.avium_104 -------------MMVLVLLA-------SGCGWKPPAPPQARPDTCKASD
MSMEG_4475|M.smegmatis_MC2_155 ------------MSVVVVSLG----FLAAGCGWGPSSAPPPPPDTCTAAD
TH_0726|M.thermoresistible__bu -----------VSLVIGLIIG-----LVAGCGWSPPSSQPPPPDTCTPTD
Mflv_2725|M.gilvum_PYR-GCK --MPISRARHGYVALTLVMLA------VSACGWSPP-APAPTSTAAECGP
Mvan_3809|M.vanbaalenii_PYR-1 --MSIPR----LVVALVTVAA------LSGCGWSPP-APPPTSTAAECGP
. ..*. .. . . :.
MMAR_3638|M.marinum_M GPTSQTVRQAIAAVPVGVPNSTWVEIARGHTKNCRLYWVQIIPTIASE-S
MUL_1475|M.ulcerans_Agy99 GPSSQTVRQAIAAVPVGVPNSTWVEIARGHTKNCRLYWVQIIPTIASE-S
Mb2357c|M.bovis_AF2122/97 GPTADTVRQAIAAVPIVVPGSKWVEITRGHTRNCRLHWVQIIPTIASQ-S
Rv2330c|M.tuberculosis_H37Rv GPTADTVRQAIAAVPIVVPGSKWVEITRGHTRNCRLHWVQIIPTIASQ-S
MAV_2076|M.avium_104 GPTADTVRQAITAVPIAIPGTIWVEIGRGHTRNCRLHWVQIIPTIASE-S
MSMEG_4475|M.smegmatis_MC2_155 GPDPMVVAQEIAKLPAPHGGSQWVQVRDGHTTDCRLHWVQVGLTDPQP-N
TH_0726|M.thermoresistible__bu GPAPDTVAAAIDQLPR-VDGQPWRETARGHTHNCRLYWVQVMPITGQDRD
Mflv_2725|M.gilvum_PYR-GCK GPTEQAVESEYALLPA----GAWRETARGSAADCRLQWVVVSSGDQPD--
Mvan_3809|M.vanbaalenii_PYR-1 GPAADVVAAEIAMLPP----AEWEETERGHSADCRLNWVVVSSG-HFD--
** .* :* * : * : :*** ** :
MMAR_3638|M.marinum_M TPQQLLFFDHNTALGSPTRNPKPYIRVLPSGQDTVTVQYQWRVGSDPQCC
MUL_1475|M.ulcerans_Agy99 APQQLLFFDHNTALGSPTRNPKPYIRVLPSGQDTVTVQYQWRVGSDPQCC
Mb2357c|M.bovis_AF2122/97 TPQQLLFFDRNIPLGSPTRNPKPYITVLPAGDDTVTVQYQWQIGSDQECC
Rv2330c|M.tuberculosis_H37Rv TPQQLLFFDRNIPLGSPTRNPKPYITVLPAGDDTVTVQYQWQIGSDQECC
MAV_2076|M.avium_104 TPQQLLFFDHNTPLGTATPNAKPYITVLPPSDDTVTVQYQWQKGNDQLCC
MSMEG_4475|M.smegmatis_MC2_155 SVGQLLFFDRQTPLGTATPEPRPYINVVNNGEDTVTVNYQWQQGEDTPEA
TH_0726|M.thermoresistible__bu TPQQVLFFDRNSPIGTATPDPRPYLLVLNSGPETVTVQYQWRQGDDEPCC
Mflv_2725|M.gilvum_PYR-GCK SPQQVLFFDDGAPVGSPTMDPRPYITVTAQGERDAVVQYQWRQGGDEPCC
Mvan_3809|M.vanbaalenii_PYR-1 APQQVLFFDGTEPVGSPTPEPRRFITVIPQGRSDAVVQYQWRQGQDEPCC
: *:**** .:*:.* :.: :: * . ..*:***: * * .
MMAR_3638|M.marinum_M PTGSGTVRFQVGPDGKLEALGPIPHQ
MUL_1475|M.ulcerans_Agy99 PTGSGTVRFQVGPDGKLKALGPIPHQ
Mb2357c|M.bovis_AF2122/97 PTGIGTVRFHIGSDGKLEALGSIPHQ
Rv2330c|M.tuberculosis_H37Rv PTGIGTVRFHIGSDGKLEALGSIPHQ
MAV_2076|M.avium_104 PTGIGTVKFRIGPDGKLQTIGKIPNQ
MSMEG_4475|M.smegmatis_MC2_155 PTGIATVRFRIGDDGRLVAVDPLPSP
TH_0726|M.thermoresistible__bu PTGIGTVRFQIDDDGRLRALDPIPGP
Mflv_2725|M.gilvum_PYR-GCK PTGIGTARVTLE-DGRLTVLDPIPNE
Mvan_3809|M.vanbaalenii_PYR-1 PTGIGTARVTLE-EGRLTILDPIPGP
*** .*.:. : :*:* :. :*