For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KPFLGRLLACLLVGCVLCPAVFGCAPTHQRGGVRTPAALLSYLGQAQIPFGASVDGAVIGGLSGISYDAD RQVYYVISDDRSDRGPGRFFTMRLALSDIGISDITVTGAYSLLDRDGKPFGSLNRSSTPPVISPDPEGIA VDTARRRLYWSSEGERLTDRQSGLLDPWIRIADLDGRYLGQFTLPPNLAVSAQRTGPRNNQALEGLALTP DGRSLYAAMEEPGLQDEPPANTGRQVLTRFTKFDVETAAPTAQYAYPMEPAASPAVRNGVSDLVALSDTT FLVVERTATVPPVIRIFRAEIGAATDVLSMPTIHGTPLTPMRKTLAADLPALAGTTAAAALSPLNNIEGI TLGPKLADGRQSVVLVSDNDFSPAKVTQLLLWAMP*
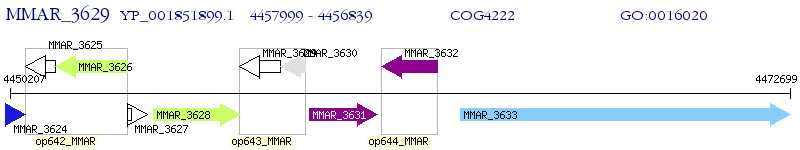
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3629 | - | - | 100% (386) | hypothetical protein MMAR_3629 |
| M. marinum M | MMAR_4416 | - | 2e-09 | 25.49% (306) | hypothetical protein MMAR_4416 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0772 | - | 6e-95 | 51.80% (361) | phytase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3629|M.marinum_M --MKPFLGRLLACLLVGCVLCPAVFGCAPTHQRGGVRTPAALLSYLGQAQ
MSMEG_0772|M.smegmatis_MC2_155 MGMNPLF-RLTRSMTAAAVMAAAAAATTP------IAVADTPLEYLGQWQ
*:*:: ** .: ...*:..*. . :* : .. : *.**** *
MMAR_3629|M.marinum_M IPFGASVDGAVIGGLSGISYDADRQVYYVISDDRSDRGPGRFFTMRLALS
MSMEG_0772|M.smegmatis_MC2_155 LPPQEAGAPNRIGGLSGITYDPAHQTYYLISDDKTEHGPVRFHTARIALS
:* : *******:**. :*.**:****::::** **.* *:***
MMAR_3629|M.marinum_M DIGISDITVTGAYSLLDRDGKPFGSLNRSSTPPVISPDPEGIAVDTARRR
MSMEG_0772|M.smegmatis_MC2_155 GNGVDDVQFVGVQPLLDESGKPFGPNNFTSNPPVLAPDAEAIAYDAGRRL
. *:.*: ..*. .***..*****. * :*.***::**.*.** *:.**
MMAR_3629|M.marinum_M LYWSSEGERLTDRQSG---LLDPWIRIADLDGRYLGQFTLPPNLAVS-AQ
MSMEG_0772|M.smegmatis_MC2_155 LYWTSEG-YIHDDASGKPAILQPWIRVAGLDGAYQGEFALPPSFMFTGDQ
***:*** : * ** :*:****:*.*** * *:*:***.: .: *
MMAR_3629|M.marinum_M RTGPRNNQALEGLALTPDGRSLYAAMEEPGLQDEPPANTGRQVLTRFTKF
MSMEG_0772|M.smegmatis_MC2_155 TAGPRPNGVLEGLSLTPDGRYLFAAMEQPLYQDGPATDENHTGLVRITKF
:*** * .****:****** *:****:* ** *.:: .: *.*:***
MMAR_3629|M.marinum_M DVETAAPTAQYAYPMEPAASPAVRNGVSDLVALSDTTFLVVERTATVP--
MSMEG_0772|M.smegmatis_MC2_155 DTVTRTPVAEYAYPVDRAAPETGFNGVSDILALSETSFLVIERAASLPRQ
*. * :*.*:****:: **. : *****::***:*:***:**:*::*
MMAR_3629|M.marinum_M PVIRIFRAEIGAATDVLSMPTIHGTPLTPMRKTLAADLPALAGTTAAAAL
MSMEG_0772|M.smegmatis_MC2_155 VAVSVYRADTGPASDVLDVASLRDQAVTPMTKTLAADLRATPG------L
.: ::**: *.*:***.:.:::. .:*** ******* * .* *
MMAR_3629|M.marinum_M SPLNNIEGITLGPKLADGRQSVVLVSDNDFSPAKVTQLLLWAMP--
MSMEG_0772|M.smegmatis_MC2_155 DRLDNIEGLTLGPRLPDGRQSVVLVSDDNFQPFEVTQFLAFALPRS
. *:****:****:*.***********::*.* :***:* :*:*