For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMAAYHCPVCDYVYDEAVGAPREGYPAGTSWADVDETWNCPDCGVREKIDFQLQEA
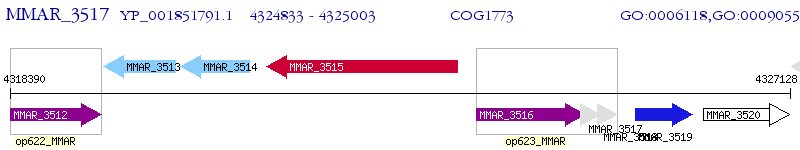
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3517 | rubA_1 | - | 100% (56) | rubredoxin RubA_1 |
| M. marinum M | MMAR_1292 | rubA | 5e-19 | 64.71% (51) | rubredoxin RubA |
| M. marinum M | MMAR_1293 | rubB | 3e-11 | 49.02% (51) | rubredoxin RubB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3279c | rubA | 1e-19 | 68.63% (51) | rubredoxin RUBA |
| M. gilvum PYR-GCK | Mflv_4720 | - | 2e-19 | 66.67% (51) | rubredoxin-type Fe(Cys)4 protein |
| M. tuberculosis H37Rv | Rv3251c | rubA | 1e-19 | 68.63% (51) | rubredoxin RUBA |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3597c | - | 2e-11 | 49.02% (51) | rubredoxin RubB |
| M. avium 104 | MAV_4214 | - | 2e-19 | 66.67% (51) | rubredoxin |
| M. smegmatis MC2 155 | MSMEG_1840 | - | 1e-20 | 70.59% (51) | rubredoxin |
| M. thermoresistible (build 8) | TH_0469 | rubA | 1e-20 | 68.63% (51) | PROBABLE RUBREDOXIN RUBA |
| M. ulcerans Agy99 | MUL_2748 | rubA_1 | 5e-32 | 100.00% (56) | rubredoxin RubA_1 |
| M. vanbaalenii PYR-1 | Mvan_1743 | - | 3e-19 | 64.71% (51) | rubredoxin-type Fe(Cys)4 protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3517|M.marinum_M ---------MMAAYHCPVCDYVYDEAVGAPREGYPAGTSWADVDETWNCP
MUL_2748|M.ulcerans_Agy99 ---------MMAAYHCPVCDYVYDEAVGAPREGYPAGTSWADVDETWNCP
Mb3279c|M.bovis_AF2122/97 ----------MAAYRCPVCDYVYDEANGDAREGFPAGTGWDQIPDDWCCP
Rv3251c|M.tuberculosis_H37Rv ----------MAAYRCPVCDYVYDEANGDAREGFPAGTGWDQIPDDWCCP
Mflv_4720|M.gilvum_PYR-GCK ----------MTAYRCPGCDYTYDESKGEPREGFPAGTRWADVPDEWCCP
Mvan_1743|M.vanbaalenii_PYR-1 ----------MTAFRCPGCDYVYDESKGEPREGFPAGTAWAEVPDEWCCP
MAV_4214|M.avium_104 ----------MTRYRCPGCDYIYDEAKGAPREGFPAGTPFADIPDDWCCP
TH_0469|M.thermoresistible__bu ----------VSAYRCPVCDYVYDEAKGAAREGFPAGTPWSEVPDDWPCP
MSMEG_1840|M.smegmatis_MC2_155 ----------MSSYRCPVCDYIYDEAKGAAREGFPPGTAWDDVPDDWCCP
MAB_3597c|M.abscessus_ATCC_199 MSDETCVDSEYKLYECMQCGFQYDEALGWPEDGIEPGTRWDDIPEDWSCP
:.* *.: ***: * ..:* .** : :: : * **
MMAR_3517|M.marinum_M DCGVREKIDFQLQEA---
MUL_2748|M.ulcerans_Agy99 DCGVREKIDFQLQEA---
Mb3279c|M.bovis_AF2122/97 DCAVREKVDFEKIGG---
Rv3251c|M.tuberculosis_H37Rv DCAVREKVDFEKIGG---
Mflv_4720|M.gilvum_PYR-GCK DCAVREKVDFEEMGVKS-
Mvan_1743|M.vanbaalenii_PYR-1 DCAVREKVDFEEMGVKR-
MAV_4214|M.avium_104 DCAVREKVDFENLIGQE-
TH_0469|M.thermoresistible__bu DCGVRDKVDFEPVTG---
MSMEG_1840|M.smegmatis_MC2_155 DCGVREKIDFEEMGVKQ-
MAB_3597c|M.abscessus_ATCC_199 DCGA-AKADFVMVEIARP
**.. * **