For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAAVVVIGLGNRYRRDDGVGVAAATALRELALSGVRVVTDLADPMSLLEAWSGAGLAVLIDAAVATPATP GRVRRCALGDLAAGSSGVSSHRADVAKAYALGQALDRVPERLVIITVDVADIGHGVGLTPQVLDAIPVVV GLAVDEIGRPRPRADV*
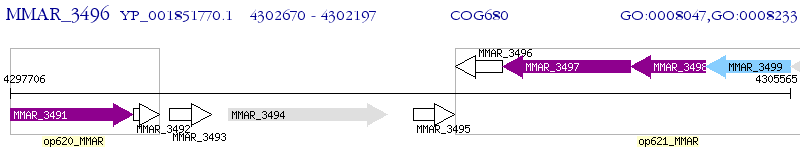
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3496 | - | - | 100% (157) | hypothetical protein MMAR_3496 |
| M. marinum M | MMAR_1864 | hyaD | 2e-06 | 29.81% (161) | nickel/iron hydrogenase maturation factor, HyaD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3927 | - | 9e-34 | 46.41% (153) | peptidase M52, hydrogen uptake protein |
| M. thermoresistible (build 8) | TH_1029 | - | 2e-09 | 32.08% (159) | peptidase M52, hydrogen uptake protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2367 | - | 2e-07 | 31.69% (142) | hydrogenase maturation protease |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1029|M.thermoresistible__bu VIPTSAEADIDIEPPGCAVLVVGCGNLLRGDDGVGPILIRHLWERGVPDG
Mvan_2367|M.vanbaalenii_PYR-1 --------------------MIGCGNLLRGDDGVGPVLVRHLWERGVPDG
MMAR_3496|M.marinum_M --------------MNAAVVVIGLGNRYRRDDGVGVAAATALRELALSGV
MSMEG_3927|M.smegmatis_MC2_155 ----------------MTVLVIGIGNEARRDDGVGIAAVHEIAQRRLPGV
::* ** * ***** : : :..
TH_1029|M.thermoresistible__bu ARLVDGGTAGMDVAFQMKGAQRVVIVDAALTG-AAPGTVYRVPGAELAEL
Mvan_2367|M.vanbaalenii_PYR-1 AKLVDGGTAGMDVAFQMNGARRVVIIDAAATG-SAPGTTFRVPGAELAEL
MMAR_3496|M.marinum_M RVVTDLADP-MSLLEAWSGAGLAVLIDAAVATPATPGRVRRCALGDLAAG
MSMEG_3927|M.smegmatis_MC2_155 EAMVTSGDP-GELLDAWAGVPTVIAVDAAVSPSATPGTVRRWTPADLPRD
:. . . .: *. .: :*** : ::** . * . .:*.
TH_1029|M.thermoresistible__bu PPLQGLHTHSFRWDHSIAFARWALGDACPDDITVFLIEAGGVELGAELSA
Mvan_2367|M.vanbaalenii_PYR-1 PPLQGLHTHSFRWDHAIAFARWALGDACPTDITVFLIEVDSVEMGADLSE
MMAR_3496|M.marinum_M --SSGVSSHRADVAKAYALG--QALDRVPERLVIITVDVADIGHGVGLTP
MSMEG_3927|M.smegmatis_MC2_155 --LGGVSSHALGLAGAYRLG--EALGQSPGRLIVLTVDVVDVGYGFGMTD
*: :* : :. . * : :: ::. .: * ::
TH_1029|M.thermoresistible__bu PVAAAMEEVIELIEAEFLAGLRPRPDGRAKVEFTADGYLRLDATLAASRF
Mvan_2367|M.vanbaalenii_PYR-1 PVATAMEEVLSIVERDYLAPLRPAGRREVTVEFTADGYLRLDAELAAARF
MMAR_3496|M.marinum_M QVLDAIPVVVGLAVDEIGR---PRPRADV---------------------
MSMEG_3927|M.smegmatis_MC2_155 AVAGAVPDVVAVILREAGL---HEAVRDV---------------------
* *: *: : : .
TH_1029|M.thermoresistible__bu PSDAVAAVRRDTELWVLPLRGPRSGGLLLKQRTPAGDRAVLVREVLNDDI
Mvan_2367|M.vanbaalenii_PYR-1 PSDAVAAVVRDGALWLIPLRGPRSGGLLLKQRTPAGDRSLLVREVLMDLA
MMAR_3496|M.marinum_M --------------------------------------------------
MSMEG_3927|M.smegmatis_MC2_155 --------------------------------------------------
TH_1029|M.thermoresistible__bu PVGVREAFWDDGRSALRIPLGSHV----
Mvan_2367|M.vanbaalenii_PYR-1 MTGVRGASWDDDQSALRIPLGDEPGLDR
MMAR_3496|M.marinum_M ----------------------------
MSMEG_3927|M.smegmatis_MC2_155 ----------------------------