For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAVAAMSWWQVISLAVVQGLTEFLPVSSSGHLAVVSRVFFSDDAGASFTAVTQLGTEAAVLVYFARDIVR ILRAWFDGLVVTSHRNADYRLGWYVIIGTIPICVMGLLFKDEIRSGVRNLWVVATALVVFSGVIALAEYL GRQSRHVEQLTWRDGLVVGVAQTLALVPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSL PDAFHPVTEGMSATGPQLLVATLIAFVVGLAAVSWFLRFLLRHSMYWFVGYRVVVGVVVLILLATGTVAA T*
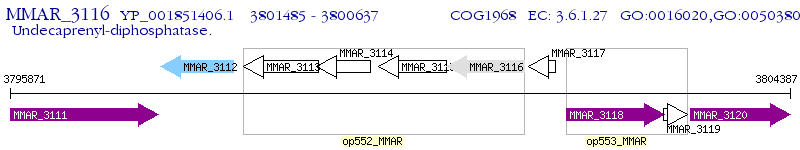
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3116 | - | - | 100% (282) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2160c | uppP | 1e-138 | 86.59% (276) | undecaprenyl pyrophosphate phosphatase |
| M. gilvum PYR-GCK | Mflv_3043 | - | 1e-125 | 80.73% (275) | undecaprenyl pyrophosphate phosphatase |
| M. tuberculosis H37Rv | Rv2136c | uppP | 1e-138 | 86.59% (276) | undecaprenyl pyrophosphate phosphatase |
| M. leprae Br4923 | MLBr_01297 | - | 1e-133 | 83.33% (282) | undecaprenyl pyrophosphate phosphatase |
| M. abscessus ATCC 19977 | MAB_2108 | - | 1e-124 | 76.87% (281) | undecaprenyl pyrophosphate phosphatase |
| M. avium 104 | MAV_2853 | - | 5e-38 | 33.55% (304) | undecaprenyl pyrophosphate phosphatase |
| M. smegmatis MC2 155 | MSMEG_4194 | - | 1e-123 | 78.55% (275) | undecaprenyl pyrophosphate phosphatase |
| M. thermoresistible (build 8) | TH_2364 | - | 1e-118 | 75.36% (276) | Possible conserved transmembrane protein |
| M. ulcerans Agy99 | MUL_2366 | - | 1e-153 | 99.28% (276) | undecaprenyl pyrophosphate phosphatase |
| M. vanbaalenii PYR-1 | Mvan_3483 | - | 1e-122 | 78.18% (275) | undecaprenyl pyrophosphate phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3116|M.marinum_M -MSAVAAMSWWQVISLAVVQGLTEFLPVSSSGHLAVVSRVF---FSDDAG
MUL_2366|M.ulcerans_Agy99 -------MSWWQVISLAVVQGLTEFLPVSSSGHLAVVSRVF---FSDDAG
Mb2160c|M.bovis_AF2122/97 -------MSWWQVIVLAAAQGLTEFLPVSSSGHLAIVSRIF---FSGDAG
Rv2136c|M.tuberculosis_H37Rv -------MSWWQVIVLAAAQGLTEFLPVSSSGHLAIVSRIF---FSGDAG
MLBr_01297|M.leprae_Br4923 -MTAVSAMSWWQVIVLAVVQGLTEFLPVSSSGHLAIVSRIL---FTGDAG
MAB_2108|M.abscessus_ATCC_1997 MDSAVAAMSWLQVIVLAVVQGLTEFLPVSSSGHLAIVSRVF---FHDDAG
Mflv_3043|M.gilvum_PYR-GCK -------MSWLQVVVLSVLQGLTEFLPVSSSGHLAIASRVF---FEDDAG
Mvan_3483|M.vanbaalenii_PYR-1 -------MSWLQVVVLSVLQGLTEFLPVSSSGHLAIASRVF---FEDDAG
MSMEG_4194|M.smegmatis_MC2_155 -------MSWLQVIVLSIVQGLTEFLPVSSSGHLAITSQVF---FDDDAG
TH_2364|M.thermoresistible__bu -------MSWLQVIVLSVVQGLTEFLPISSSGHLAITSKVF---FDGDAG
MAV_2853|M.avium_104 ---MTAQLSYVEAVVVGAFQGVTELFPVSSLGHAVLVPALVGGRWAQDLS
:*: :.: :. **:**::*:** ** .:.. :. : * .
MMAR_3116|M.marinum_M AS--------FTAVTQLGTEAAVLVYFARDIVRILRAWFDGLVVTSHRNA
MUL_2366|M.ulcerans_Agy99 AS--------FTAVTQLGTEAAVLVYFARDIVRILRAWFDGLVVKSHRNA
Mb2160c|M.bovis_AF2122/97 AS--------FTAVSQLGTEAAVVIYFARDIVRILSAWVHGLVVKAHRNT
Rv2136c|M.tuberculosis_H37Rv AS--------FTAVSQLGTEAAVVIYFARDIVRILSAWLHGLVVKAHRNT
MLBr_01297|M.leprae_Br4923 AS--------FTAVSQLGTEVAVLVYFGRDIVRILHAWCRGLTVTLHRTA
MAB_2108|M.abscessus_ATCC_1997 AS--------FTAVTQLGTEAAVLLYFAKDIWRILRAWFDGLFVKAHRSF
Mflv_3043|M.gilvum_PYR-GCK AS--------FTAVCQLGTEAAVLVYFARDIARILKAWFAGLFGSGERSP
Mvan_3483|M.vanbaalenii_PYR-1 AS--------FTAVSQLGTEVAVLVYFARDIVRIVKAWFAGLFRAGQRSA
MSMEG_4194|M.smegmatis_MC2_155 AS--------FTAVTQLGTEFAVLIYFAKDIGRIIKSWFLGLRAPEHRDA
TH_2364|M.thermoresistible__bu AS--------FTAVTQIGTEAAVLLYFARDIGRIVKAWFGGLFDAARRDA
MAV_2853|M.avium_104 VSAHRSPYLAFIVGLHVATAAALLVFFWRDWVRILAGFFSSLRHRRIRTP
.* * . ::.* *::::* :* **: .: .* *
MMAR_3116|M.marinum_M DYRLGWYVIIGTIPICVMGLLFKDEIRSGVRNLWVVATALVVFSGVIALA
MUL_2366|M.ulcerans_Agy99 DYRLGWYVIIGTIPICVLGLLFKDEIRSGVRNLWVVATALVVFSGVIALA
Mb2160c|M.bovis_AF2122/97 DYRLGWYVIIGTIPICILGLFFKDDIRSGVRNLWVVVTALVVFSGVIALA
Rv2136c|M.tuberculosis_H37Rv DYRLGWYVIIGTIPICILGLFFKDDIRSGVRNLWVVVTALVVFSGVIALA
MLBr_01297|M.leprae_Br4923 DYWLGWYVIIGTIPICILGLVCKDEIRSGIRPLWVVATALVAFSGVIAFA
MAB_2108|M.abscessus_ATCC_1997 DYWMGWYVIVGTMPIGILGLAFKDQIRSGARNLWLISASLIIFALVIAAA
Mflv_3043|M.gilvum_PYR-GCK DYWLGWWVIIGTIPISVIGLLFKDEIRTGARNLWLVATAMIVFSFVIAAA
Mvan_3483|M.vanbaalenii_PYR-1 DYWLGWWVIIGTIPISVVGLLFKDEIRTGARNLWLVATAMIVFSFVIAAA
MSMEG_4194|M.smegmatis_MC2_155 DYRLGWFVIVGTIPIGVFGLLFKDEIRTGARNLWLVATALIVFSVVIAAA
TH_2364|M.thermoresistible__bu DYRMGWFVIIGSIPIGVFGLVFTDDIRTGARNLWLVATALIVFSVVIAAA
MAV_2853|M.avium_104 DERLAWLIVVGTIPVGLAGLALEQLFRTTLGKPVPAAAFLLLNSVALYAG
* :.* :::*::*: : ** : :*: : :: : .: .
MMAR_3116|M.marinum_M EYLGRQSR----------------------HVEQLTWRDGLVVGVAQTLA
MUL_2366|M.ulcerans_Agy99 EYLGRQSR----------------------HVEQLTWRDGLVVGVAQTLA
Mb2160c|M.bovis_AF2122/97 EYVGRQSR----------------------HIERLTWRDAVVVGIAQTLA
Rv2136c|M.tuberculosis_H37Rv EYVGRQSR----------------------HIERLTWRDAVVVGIAQTLA
MLBr_01297|M.leprae_Br4923 EYVGRQNR----------------------CIEQLNWRDALVVGVAQTLA
MAB_2108|M.abscessus_ATCC_1997 EYYGRQVR----------------------HTEQLTMKDAVIVGSAQALA
Mflv_3043|M.gilvum_PYR-GCK EYAGRQTR----------------------RVEQLTWRDSVIVGFAQCLA
Mvan_3483|M.vanbaalenii_PYR-1 EYYGRQAR----------------------RVEQLTWRDSIIVGLAQCLA
MSMEG_4194|M.smegmatis_MC2_155 EYYGRQVR----------------------QVEQLTWRDSVIVGLAQCLA
TH_2364|M.thermoresistible__bu EYLGRQNR----------------------QREELTWKDAVIVGFAQCLA
MAV_2853|M.avium_104 EVLRRRVAPVADEPAVPDAEQQHGDEASDNRLAQLPLRRGVLIGAAQILA
* *: .* : .:::* ** **
MMAR_3116|M.marinum_M LVPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSLPDAFH
MUL_2366|M.ulcerans_Agy99 LVPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSLPDAFH
Mb2160c|M.bovis_AF2122/97 LVPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSLPDAFH
Rv2136c|M.tuberculosis_H37Rv LVPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSLPDAFH
MLBr_01297|M.leprae_Br4923 LIPGVSRSGSTISAGLFLGLDRELAARFGFLLAIPAVFASGLFSIPDAFH
MAB_2108|M.abscessus_ATCC_1997 LIPGVSRSGATISAGLFLGLDRPASARFGFLLAIPAVLASGLFSLPDAFH
Mflv_3043|M.gilvum_PYR-GCK LVPGVSRSGATISAGLFLGMDRELAARFGFLLAIPAVFASGLFSLPDAFA
Mvan_3483|M.vanbaalenii_PYR-1 LVPGVSRSGATISAGLFLGMHRELAARFGFLLAIPAVFASGLFSLPDAFE
MSMEG_4194|M.smegmatis_MC2_155 LMPGVSRSGATISAGLFLGLKREVAARFGFLLAIPAVLASGLFSLPDAFH
TH_2364|M.thermoresistible__bu LIPGVSRSGATISAGLFLGQNRALAARFGFLLAIPAVFASGLFSLPDAFN
MAV_2853|M.avium_104 LLPGISRSGITIVAGLWRGLSHEDAARFSFLLATPIILAAGVYKIPELFG
*:**:**** ** ***: * : :***.**** * ::*:*::.:*: *
MMAR_3116|M.marinum_M PVTEGMSATGPQLLVATLIAFVVGLAAVSWFLRFLLRHSMYWFVGYRVVV
MUL_2366|M.ulcerans_Agy99 PVTEGMSATGPQLLVATLIAFVVGLAAVSWFLRFLLRHSMYWFVGYRVVV
Mb2160c|M.bovis_AF2122/97 PVTEGMSATGPQLLVATLIAFVLGLTAVAWLLRFLVRHNMYWFVGYRVLV
Rv2136c|M.tuberculosis_H37Rv PVTEGMSATGPQLLVATLIAFVLGLTAVAWLLRFLVRHNMYWFVGYRVLV
MLBr_01297|M.leprae_Br4923 PITEGMSATGAQLLVATVIAFVVGLVAVSWLLRFLVQHNLYWFVGYRIVV
MAB_2108|M.abscessus_ATCC_1997 PVTEGMSATGPQLLVATLIAFVIGYAAVAWLLRFISNHSMYWFVGYRVIL
Mflv_3043|M.gilvum_PYR-GCK PVGEGMSATGPQLLVSTVIAFVVGYAAVAWFLRFLVRHSMYWFVGYRIIL
Mvan_3483|M.vanbaalenii_PYR-1 PVGEGMSASGAQLFVSIVIAFVVGYAAVAWFLRFLVRHSMYWFVGYRIVL
MSMEG_4194|M.smegmatis_MC2_155 PVGEGMSASGPQLIVATVIAFVVGFAAIAWFLKFLVSHSMYWFVGYRVVL
TH_2364|M.thermoresistible__bu PSGAGMSATGAQLLVATVIAFVVGFAAVAWFLRFLVRHSMYWFVGYRMLL
MAV_2853|M.avium_104 PLGAGIGG---QVLAGSIASFVCAYLAVRYLTRYFQTRTLTPFAIYCAVA
* *:.. *::.. : :** . *: :: ::: :.: *. * :
MMAR_3116|M.marinum_M GVVVLILLATGTVAAT
MUL_2366|M.ulcerans_Agy99 GVVVLILLATGTVAAT
Mb2160c|M.bovis_AF2122/97 GTGMLVLLATGTVAAT
Rv2136c|M.tuberculosis_H37Rv GTGMLVLLATGTVAAT
MLBr_01297|M.leprae_Br4923 GVGVLILLAVKTVAAT
MAB_2108|M.abscessus_ATCC_1997 GLAVMGLLAAGVVSAS
Mflv_3043|M.gilvum_PYR-GCK GSVVLILLSTGVVAAI
Mvan_3483|M.vanbaalenii_PYR-1 GTVVLVLLSAGVVSAI
MSMEG_4194|M.smegmatis_MC2_155 GVVVLALLGTGVLAAQ
TH_2364|M.thermoresistible__bu GAVVLVLLGAGVVTAT
MAV_2853|M.avium_104 GGASLVWLALR-----
* : *.