For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AELSLISVGDTVRGMWHALSRRDWDGVKTFLSPDCLYVDMPVPALSARGPDDIVTRLKLGLEPLAGYENH EGLLLSSGSDVMYEHSETWTFATGEQGLLRFVTVHQVLDGKITVWKDYWDFNSLVAFAPPQHFENLGEAD QGWIFDASSLV*
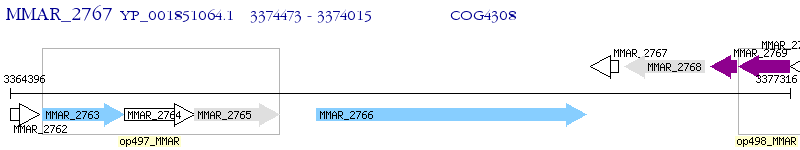
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2767 | - | - | 100% (152) | hypothetical protein MMAR_2767 |
| M. marinum M | MMAR_1973 | - | 8e-05 | 26.96% (115) | hypothetical protein MMAR_1973 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3314 | - | 2e-52 | 63.38% (142) | limonene-1,2-epoxide hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2596c | - | 2e-47 | 59.71% (139) | hypothetical protein MAB_2596c |
| M. avium 104 | MAV_2823 | - | 1e-68 | 76.87% (147) | limonene-1,2-epoxide hydrolase catalytic domain-containing |
| M. smegmatis MC2 155 | MSMEG_2698 | - | 2e-06 | 27.83% (115) | hypothetical protein MSMEG_2698 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2980 | - | 3e-39 | 95.89% (73) | hypothetical protein MUL_2980 |
| M. vanbaalenii PYR-1 | Mvan_3036 | - | 1e-49 | 60.84% (143) | limonene-1,2-epoxide hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2767|M.marinum_M -----MAELS---LISVGDTVRG----MWHALSRRD--WDGVKTFLS---
MAV_2823|M.avium_104 -------------MIAVEDVVRG----LWQALSRRD--WDAVKTFLS---
Mflv_3314|M.gilvum_PYR-GCK --MPVDQIAR---EAPVEATVYG----MWAALSARD--WDAVKTFLA---
Mvan_3036|M.vanbaalenii_PYR-1 --MPVDQVAN---TGQAEETVQA----MWEALSARD--WTALKTFLS---
MAB_2596c|M.abscessus_ATCC_199 -------------MAAQASLALG----LWLALAQRN--WEAVKTFLD---
MUL_2980|M.ulcerans_Agy99 MRAPIGIGAARWLTAQEGPSVIGRYGRTFFDFGWRYGAWDVARAVSEGLG
MSMEG_2698|M.smegmatis_MC2_155 -------------MQNPIATVET----FLNALQEQD--FDTAENALA---
. : : : .
MMAR_2767|M.marinum_M --PDCLYVDMPV-PALSARGPDDIVTRLKLGLEPLAG----YENHEGLLL
MAV_2823|M.avium_104 --DDCLYVDMPV-PALSARGPEDIVKRLKMGLEQLAG----YQNHPGVLV
Mflv_3314|M.gilvum_PYR-GCK --PDCIYLDMPVGPAAAARGPEDIVKRLKIGIEPLAS----YENFTGLMV
Mvan_3036|M.vanbaalenii_PYR-1 --ADCIYLDVPVGPAAAARGPEDIVKRLKIGLEPLAA----YRNFPGLMV
MAB_2596c|M.abscessus_ATCC_199 --DGCIYLDMPVGPAAAARGPENIVKRLKMGLEPLKD----YAHHDGLII
MUL_2980|M.ulcerans_Agy99 WRQDVFVAGLPVCGHAGSRAVGARSGRHRHAAEARVGTPGRLREPRRTAA
MSMEG_2698|M.smegmatis_MC2_155 --QNLVYQNVGMPTIYGR----NRAMKLFRSMQGRAG----FEVKIHRSA
. . .: : . : . :
MMAR_2767|M.marinum_M SSGSDVMYEHSETWTFATGEQGLLRFVTVHQVLDGKITVWKDYWDFNSLV
MAV_2823|M.avium_104 SNGSDVLYEHSETWTFATGEQGVLRFVTVHKVIDGKVTVWKDYWDMQSLV
Mflv_3314|M.gilvum_PYR-GCK SNGADVMYEHHEEWHWDTGESAVLQFVTVHRVEDGRITLWKDYWDMAALA
Mvan_3036|M.vanbaalenii_PYR-1 SNGADVMYEHSEQWHWATGESALLKFVTVHLVEGGEITLWKDYWDMGALA
MAB_2596c|M.abscessus_ATCC_199 SNGQDVMYEHSETWTWDTGETALLHFASVLRVDDGKITLWKDYWDMGALT
MUL_2980|M.ulcerans_Agy99 VQRSDVMYEHSETWTFATGEQGLLRFVTVHQVVDGKITVWKDYWDFNSLA
MSMEG_2698|M.smegmatis_MC2_155 ADGAAVLNERTDVLTFGP-LRVQFWVCGVFEVHDGRITLWRDYFDYLDIF
. *: *: : : . : . * * .*.:*:*:**:* :
MMAR_2767|M.marinum_M AFAPPQHFENLGEADQGWIFDASSLV
MAV_2823|M.avium_104 AFAPPNHFDGLANGDTSWVFDASALV
Mflv_3314|M.gilvum_PYR-GCK GKAPPTWLADFAEADMSWVFDATGLV
Mvan_3036|M.vanbaalenii_PYR-1 DHAPPTWLADFAGADMSWVFDATGLV
MAB_2596c|M.abscessus_ATCC_199 NSAPPSWMENLMSADMSWIYDATGQV
MUL_2980|M.ulcerans_Agy99 AFAPPQHFENLGSADQGWIFDASSLV
MSMEG_2698|M.smegmatis_MC2_155 KATVRGLLGMVVPSLRAKL-------
: : . . . :