For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQGDPDVLRLLNEQLTSELTAINQYFLHSKMQENWGFTELAAHTRDESFDEMRHAEAITDRILLLDGLPN YQRIGSLRVGQTLREQFEADLAIEYEVVNRLKPGIIMCREKQDSTSAVLFESIVADEEKHIDYLETQLEL MNKLGEELYSAQCVSRPPS
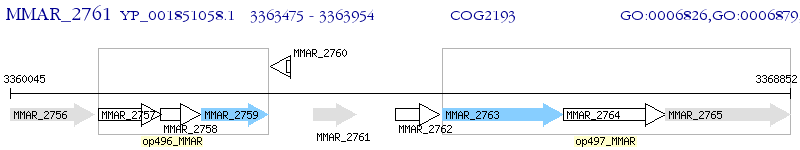
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2761 | bfrA | - | 100% (159) | bacterioferritin BfrA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1907 | bfrA | 1e-82 | 91.82% (159) | bacterioferritin |
| M. gilvum PYR-GCK | Mflv_3322 | - | 1e-72 | 83.54% (158) | bacterioferritin |
| M. tuberculosis H37Rv | Rv1876 | bfrA | 1e-82 | 91.82% (159) | bacterioferritin |
| M. leprae Br4923 | MLBr_02038 | bfrA | 1e-81 | 93.08% (159) | bacterioferritin |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_2833 | bfr | 6e-80 | 89.94% (159) | bacterioferritin |
| M. smegmatis MC2 155 | MSMEG_3564 | bfr | 4e-76 | 85.44% (158) | bacterioferritin |
| M. thermoresistible (build 8) | TH_0412 | - | 5e-73 | 82.91% (158) | PROBABLE BACTERIOFERRITIN BFRA |
| M. ulcerans Agy99 | MUL_2993 | bfrA | 5e-86 | 98.11% (159) | bacterioferritin BfrA |
| M. vanbaalenii PYR-1 | Mvan_3043 | - | 4e-76 | 86.79% (159) | bacterioferritin |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1907|M.bovis_AF2122/97 MQGDPDVLRLLNEQLTSELTAINQYFLHSKMQDNWGFTELAAHTRAESFD
Rv1876|M.tuberculosis_H37Rv MQGDPDVLRLLNEQLTSELTAINQYFLHSKMQDNWGFTELAAHTRAESFD
MMAR_2761|M.marinum_M MQGDPDVLRLLNEQLTSELTAINQYFLHSKMQENWGFTELAAHTRDESFD
MUL_2993|M.ulcerans_Agy99 MQGDLDVLRLLNEQLTSELTAINQYFLHSKMQENWGFTELAAHTRDESFD
MLBr_02038|M.leprae_Br4923 MQGDPDVLRLLNEQLTSELTAINQYFLHSKMQENWGFTELAERTRVESFD
MAV_2833|M.avium_104 MQGDPEVLRLLNEQLTSELTAINQYFLHSKMQDNWGFTELAEHTRAESFD
MSMEG_3564|M.smegmatis_MC2_155 MQGDPDVLKLLNEQLTSELTAINQYFLHSKMQDNWGFTELAEHTRAESFE
Mflv_3322|M.gilvum_PYR-GCK MQGDPEVLKLLNEQLTSELTTINQYFLHSKMQDNWGFTELAEHTRKESFE
Mvan_3043|M.vanbaalenii_PYR-1 MQGDPEVLKLLNEQLTSELTAINQYFLHSKMQDNWGFTELAEHTRKESFE
TH_0412|M.thermoresistible__bu MQGDPEILKLLNEQLTSELTAINQYFLHSKMQESWGFTELAEITRRESFD
**** ::*:***********:***********:.******* ** ***:
Mb1907|M.bovis_AF2122/97 EMRHAEEITDRILLLDGLPNYQRIGSLRIGQTLREQFEADLAIEYDVLNR
Rv1876|M.tuberculosis_H37Rv EMRHAEEITDRILLLDGLPNYQRIGSLRIGQTLREQFEADLAIEYDVLNR
MMAR_2761|M.marinum_M EMRHAEAITDRILLLDGLPNYQRIGSLRVGQTLREQFEADLAIEYEVVNR
MUL_2993|M.ulcerans_Agy99 EMRHAEAITDRILLLDGLPNYQRIGPLRVGQTLREQFEADLAIEYEVVNR
MLBr_02038|M.leprae_Br4923 EMRHAEAITDRILLLDGLPNYQRIGSLRVGQTLREQFEADLAIEYEVMSR
MAV_2833|M.avium_104 EMRHAEAITDRILLLDGLPNYQRLFSLRIGQTLREQFEADLAIEYEVMDR
MSMEG_3564|M.smegmatis_MC2_155 EMRHAETITDRILLLDGLPNYQRLFSLRVGQTLREQFEADLAIEYEVLER
Mflv_3322|M.gilvum_PYR-GCK EMVHAEEITDRILILDGLPNYQRLFSLRVGQTVREQFEADLEIEKEVVAR
Mvan_3043|M.vanbaalenii_PYR-1 EMVHAEEITDRILLLDGLPNYQRLFSLRVGQTVREQFEADLAIEYEVVAR
TH_0412|M.thermoresistible__bu EMNHAEEITDRILLLDGLPNYQRLFSLRVGQNVREQFEADLALEYEVVAR
** *** ******:*********: .**:**.:******** :* :*: *
Mb1907|M.bovis_AF2122/97 LKPGIVMCREKQDTTSAVLLEKIVADEEEHIDYLETQLELMDKLGEELYS
Rv1876|M.tuberculosis_H37Rv LKPGIVMCREKQDTTSAVLLEKIVADEEEHIDYLETQLELMDKLGEELYS
MMAR_2761|M.marinum_M LKPGIIMCREKQDSTSAVLFESIVADEEKHIDYLETQLELMNKLGEELYS
MUL_2993|M.ulcerans_Agy99 LKPGIIMCREKQDSTSAVLFESIVADEEKHIDYLETQLELMNKLGEELYS
MLBr_02038|M.leprae_Br4923 LKPGIIMCREKQDSTSAVLLEKIVADEEEHIDYLETQLALMGQLGEELYS
MAV_2833|M.avium_104 LKPAIILCREKQDSTTATLFEQIVADEEKHIDYLETQLELMDKLGVELYS
MSMEG_3564|M.smegmatis_MC2_155 LKPGIVLCREKQDATSARLLEQILADEETHIDYLETQLQLMDKLGDALYA
Mflv_3322|M.gilvum_PYR-GCK LKPGIIMCREKGDATSANLFEKILADEELHIDYLETQLDLMNKLGEGLYL
Mvan_3043|M.vanbaalenii_PYR-1 LKPGIILCREKGDATSANLFEKILADEEQHIDYLETQLELMGKLGEELYL
TH_0412|M.thermoresistible__bu LRPGIIMCREKEDATTANLFEKILADEEEHIDYLETQLELMDKLGEELYL
*:*.*::**** *:*:* *:*.*:**** ********* **.:** **
Mb1907|M.bovis_AF2122/97 AQCVSRPPT------
Rv1876|M.tuberculosis_H37Rv AQCVSRPPT------
MMAR_2761|M.marinum_M AQCVSRPPS------
MUL_2993|M.ulcerans_Agy99 AQCVSHPPS------
MLBr_02038|M.leprae_Br4923 AQCVSRPPS------
MAV_2833|M.avium_104 AQCVSRPPS------
MSMEG_3564|M.smegmatis_MC2_155 AQCVSRPPGSA----
Mflv_3322|M.gilvum_PYR-GCK SQCVSRPPQ------
Mvan_3043|M.vanbaalenii_PYR-1 AQCVSRPPS------
TH_0412|M.thermoresistible__bu AQCVARAPKNTSADG
:***::.*