For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTLNDAVGLATADRGLAVVSTVRADGTVQASLVNVGLLMHPDHGRPALGFATYGKVKLANLRARPQLAV TFRNGWRWATVEGGAELAGPDDSQPWLADREQLRLLLRDVFISAGGTHDNWDEYDRVMAEERRTVVLIEP TRIYGNG
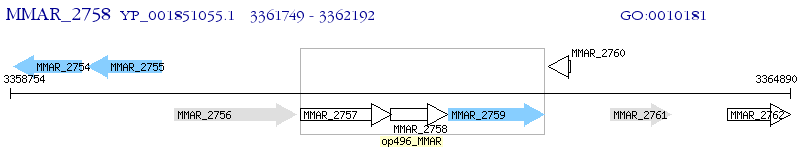
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2758 | - | - | 100% (147) | hypothetical protein MMAR_2758 |
| M. marinum M | MMAR_4298 | - | 1e-08 | 29.05% (148) | hypothetical protein MMAR_4298 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1906 | - | 7e-66 | 79.59% (147) | hypothetical protein Mb1906 |
| M. gilvum PYR-GCK | Mflv_2590 | - | 3e-59 | 70.75% (147) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. tuberculosis H37Rv | Rv1875 | - | 7e-66 | 79.59% (147) | hypothetical protein Rv1875 |
| M. leprae Br4923 | MLBr_01508 | - | 9e-10 | 30.61% (147) | hypothetical protein MLBr_01508 |
| M. abscessus ATCC 19977 | MAB_2542 | - | 7e-38 | 55.80% (138) | hypothetical protein MAB_2542 |
| M. avium 104 | MAV_2836 | - | 1e-64 | 77.55% (147) | pyridoxamine 5'-phosphate oxidase family protein |
| M. smegmatis MC2 155 | MSMEG_6576 | - | 9e-14 | 35.51% (138) | pyridoxamine 5'-phosphate oxidase family protein |
| M. thermoresistible (build 8) | TH_0458 | - | 3e-10 | 30.07% (143) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2996 | - | 5e-83 | 100.00% (147) | hypothetical protein MUL_2996 |
| M. vanbaalenii PYR-1 | Mvan_4038 | - | 2e-57 | 70.07% (147) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2590|M.gilvum_PYR-GCK ------------------MTTLHDAYALARNDNGLAVVSTLRADLTIQSS
Mvan_4038|M.vanbaalenii_PYR-1 ------------------MTPLEDAFALAVNDNGLAVVSTLRADLTIQSS
MMAR_2758|M.marinum_M ------------------MTTLNDAVGLATADRGLAVVSTVRADGTVQAS
MUL_2996|M.ulcerans_Agy99 ------------------MTTLNDAVGLATADRGLAVVSTVRADGTVQAS
Mb1906|M.bovis_AF2122/97 ------------------MTTLNEAAALAAAERGLAVVSTVRADGTVQAS
Rv1875|M.tuberculosis_H37Rv ------------------MTTLNEAAALAAAERGLAVVSTVRADGTVQAS
MAV_2836|M.avium_104 ------------------MTTLDDAVALAAAENGLAVISTVRADQTVQAS
MAB_2542|M.abscessus_ATCC_1997 ------------------MTGISAFSDLVARDHGLCVLSTVRRDGSVQSS
MLBr_01508|M.leprae_Br4923 MRFGASLLRVRRRLYGMGSQVFDDKLLAVISGNSIGVLATIKRDG--RPQ
TH_0458|M.thermoresistible__bu ----------------MTRRVFDDKLLAVIAGNSLGVLATIKRDG--RPQ
MSMEG_6576|M.smegmatis_MC2_155 -------------MTATSSDAFDPR--SLLAEARIGVLATIKANG--LPQ
: : *::*:: : ..
Mflv_2590|M.gilvum_PYR-GCK LINAGPLPHPGDGNPVLGFVTYG-KVKLANLRARPQIAVTFQ--RGWQWA
Mvan_4038|M.vanbaalenii_PYR-1 LINAGPLPHPATGLPVLGFVTYG-RVKLANLQARPQIAVTFR--RGWEWA
MMAR_2758|M.marinum_M LVNVGLLMHPDHGRPALGFATYG-KVKLANLRARPQLAVTFR--NGWRWA
MUL_2996|M.ulcerans_Agy99 LVNVGLLMHPDHGRPALGFATYG-KVKLANLRARPQLAVTFR--NGWRWA
Mb1906|M.bovis_AF2122/97 LVNVGLLPHPVSGEPSLGFTTYG-KVKLGNLRARPQLAVTFR--NGWQWA
Rv1875|M.tuberculosis_H37Rv LVNVGLLPHPVSGEPSLGFTTYG-KVKLGNLRARPQLAVTFR--NGWQWA
MAV_2836|M.avium_104 LVNVGRLAHPANGQPVLGFTTYG-KVKRANLRARPQLAVTFR--NGWQWA
MAB_2542|M.abscessus_ATCC_1997 VINAGVMPHPRTGDPIVALVAAGGTRKLDHLRTDPRTTIVVR--AGWQWT
MLBr_01508|M.leprae_Br4923 LSNVQYHFDPRDLAVRVSITEPG--VKTRNLRRDPRASILVDVDDGWSYA
TH_0458|M.thermoresistible__bu LSNVTYYFDPRRLTIQVSVTEPR--AKTRNLRRDPRASIHVSSDDGWSYA
MSMEG_6576|M.smegmatis_MC2_155 LSPVTPYYDRAADIIYVSMTDGR--AKTANLRRDPRAALEVTRADGWAWA
: . . :... * :*: *: :: . ** ::
Mflv_2590|M.gilvum_PYR-GCK TVEGRAELAGPDDPQPWLGSDYELALLLRDIFTAAGGTHDNWDEYDATMR
Mvan_4038|M.vanbaalenii_PYR-1 TVEGRAELAGPDHPQPWLGSDDTLPRLLRDIFTAAGGTHDNWDEYDRTMR
MMAR_2758|M.marinum_M TVEGGAELAGPDDSQPWLADREQLRLLLRDVFISAGGTHDNWDEYDRVMA
MUL_2996|M.ulcerans_Agy99 TVEGGAELAGPDDSQPWLADREQLRLLLRDVFISAGGTHDNWDEYDRVMA
Mb1906|M.bovis_AF2122/97 TVEGRAQLVGPDDPRPWLVDGERLRLLLREVFTAAGGTHDDWDEYDRVMA
Rv1875|M.tuberculosis_H37Rv TVEGRAQLVGPDDPRPWLVDGERLRLLLREVFTAAGGTHDDWDEYDRVMA
MAV_2836|M.avium_104 TVEGRAELVGPDDAQPWLTDADRLRLLLREVFTAAGGSHDDWDEYDRVMA
MAB_2542|M.abscessus_ATCC_1997 TVEGIAEIIGPDDPAPGV-DAESLRLLLRRIFESAGGTHDDWDTYDRVMA
MLBr_01508|M.leprae_Br4923 VAEGTAELTPPAAA-----PDDDTVEALIVLYRNIVGEHLDWDEYRQAMV
TH_0458|M.thermoresistible__bu VAEGTVTLTPEAAD-----PYDDTVEGLIELYRNIAGEHPDWDDYRRAMV
MSMEG_6576|M.smegmatis_MC2_155 TAEGTVTLTGPGTD-----PHGPEVEALVEYYRKAAGEHPDWDEYRSVMV
..** . : * : * * :** * .*
Mflv_2590|M.gilvum_PYR-GCK EQRRVAVLVTPTRVYGNG----
Mvan_4038|M.vanbaalenii_PYR-1 EQRRTAVLVTPTRIYGNG----
MMAR_2758|M.marinum_M EERRTVVLIEPTRIYGNG----
MUL_2996|M.ulcerans_Agy99 EERRTVVLIEPTRIYGNG----
Mb1906|M.bovis_AF2122/97 QEQRAVVLITPTRIYSNG----
Rv1875|M.tuberculosis_H37Rv QEQRAVVLITPTRIYSNG----
MAV_2836|M.avium_104 RERRAVVLIAPTRVYSNG----
MAB_2542|M.abscessus_ATCC_1997 AERRAAVLITPRRIYSNPAES-
MLBr_01508|M.leprae_Br4923 TDRRVLLTLPILHVYGLPPGIR
TH_0458|M.thermoresistible__bu EDRRVLLTMPISHVYGMPPGMR
MSMEG_6576|M.smegmatis_MC2_155 SDRRVLMKLSVQRVYGER--LR
::*. : : ::*.