For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTQDGTWSDESDWELDDADLADLAEAAPAPVLAIVGRPNVGKSTLVNRILGRREAVVQDIPGVTRDRVSY DALWNGRRFVVQDTGGWEPDAKGLQQLVAEQASVAMRTADAVVLVVDATVGATTADEAAARILLRSGKPV FLAANKVDSDKAEADAATLWSMGLGEPHAISAMHGRGVADLLDTVLKKLPEVAESVSAGGGPRRVALVGK PNVGKSSLLNKLAGDERSVVHDVAGTTVDPVDSLIELGGKVWRFVDTAGLRRKVGQASGHEFYASVRTRG AIDAAEVVVLLIDASQPLTEQDLRVLSMVIEAGRAVVLAYNKWDLVDEDRRDLLDREIDRELVQIRWAQR VNISAKTGRAVQKLVPAMETALASWDTRIPTGPLNSWIKEVVAATPPPVRGGKQPRILFATQATARPPTF VLFTTGFLEAGYRRFLERRLRETFGFEGSPIRINVRVREKRGAKRPGRR
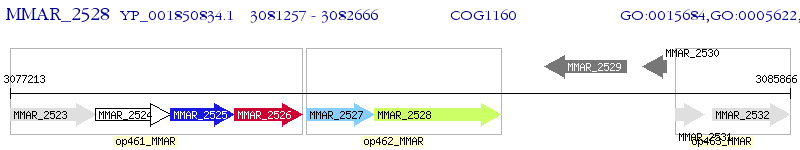
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2528 | engA | - | 100% (469) | GTP-binding protein, EngA |
| M. marinum M | MMAR_3674 | era | 1e-06 | 26.40% (197) | GTP-binding protein Era |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1740 | engA | 0.0 | 87.74% (465) | GTP-binding protein EngA |
| M. gilvum PYR-GCK | Mflv_3493 | engA | 0.0 | 83.08% (467) | GTP-binding protein EngA |
| M. tuberculosis H37Rv | Rv1713 | engA | 0.0 | 87.74% (465) | GTP-binding protein EngA |
| M. leprae Br4923 | MLBr_01372 | engA | 0.0 | 86.67% (465) | GTP-binding protein EngA |
| M. abscessus ATCC 19977 | MAB_2372 | engA | 0.0 | 79.09% (464) | GTP-binding protein EngA |
| M. avium 104 | MAV_3063 | engA | 0.0 | 89.03% (465) | GTP-binding protein EngA |
| M. smegmatis MC2 155 | MSMEG_3738 | engA | 0.0 | 84.58% (467) | GTP-binding protein EngA |
| M. thermoresistible (build 8) | TH_0203 | - | 1e-120 | 82.54% (252) | PROBABLE GTP-BINDING PROTEIN ENGA |
| M. ulcerans Agy99 | MUL_3232 | engA | 0.0 | 99.79% (469) | GTP-binding protein EngA |
| M. vanbaalenii PYR-1 | Mvan_3274 | engA | 0.0 | 83.30% (467) | GTP-binding protein EngA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3493|M.gilvum_PYR-GCK --MTGEDDQSGGDGVWFDEGDWELGADADEVAAAVEEASAPPPVLAVVGR
Mvan_3274|M.vanbaalenii_PYR-1 --MT-------DDGTWSDESDWELGP--DDISDAIEEVSAPPPVVAVVGR
MMAR_2528|M.marinum_M --MT-------QDGTWSDESDWELDDADLADLAEAAPA----PVLAIVGR
MUL_3232|M.ulcerans_Agy99 --MT-------QDGTWSDESDWELDDADLADLAEAAPA----PVLAIVGR
Mb1740|M.bovis_AF2122/97 --MT-------QDGTWVDESDWQLDDS---EIAESGAA----PVVAVVGR
Rv1713|M.tuberculosis_H37Rv --MT-------QDGTWVDESDWQLDDS---EIAESGAA----PVVAVVGR
MLBr_01372|M.leprae_Br4923 -MVS-------QDGTWSDESDWELDDS---DLAEFG------PVVAVVGR
MAV_3063|M.avium_104 --MT-------HDGTWSDESDWEAVEFELDEAAQAPPA----PVVAVVGR
MSMEG_3738|M.smegmatis_MC2_155 MSTD-------SDGTWSDESEWDLDGEFADAIHEAEDAAAPPPVLAIVGR
TH_0203|M.thermoresistible__bu --------------------------------------------------
MAB_2372|M.abscessus_ATCC_1997 MT----------DGTWEDESEWELTEFDGDEGLDEPSVP--PAVVAVVGR
Mflv_3493|M.gilvum_PYR-GCK PNVGKSTLVNRILGRREAVVQDIPGVTRDRVSYDASWLGQKFVVQDTGGW
Mvan_3274|M.vanbaalenii_PYR-1 PNVGKSTLVNRILGRREAVVQDVPGVTRDRVSYDASWSGQRFMVQDTGGW
MMAR_2528|M.marinum_M PNVGKSTLVNRILGRREAVVQDIPGVTRDRVSYDALWNGRRFVVQDTGGW
MUL_3232|M.ulcerans_Agy99 PNVGKSTLVNRILGRREAVVQDIPGVTRDRVSYDALWNGRRFVVQDTGGW
Mb1740|M.bovis_AF2122/97 PNVGKSTLVNRILGRREAVVQDIPGVTRDRVCYDALWTGRRFVVQDTGGW
Rv1713|M.tuberculosis_H37Rv PNVGKSTLVNRILGRREAVVQDIPGVTRDRVCYDALWTGRRFVVQDTGGW
MLBr_01372|M.leprae_Br4923 PNVGKSTLVNRILGRREAVVQDVPGVTRDRVSYDAMWTGRRFVVQDTGGW
MAV_3063|M.avium_104 PNVGKSTLVNRILGRREAVVQDVPGVTRDRVSYDALWTGRRFVVQDTGGW
MSMEG_3738|M.smegmatis_MC2_155 PNVGKSTLVNRILGRREAVVQDVPGVTRDRVSYDANWLGRRFVVQDTGGW
TH_0203|M.thermoresistible__bu --------------------------------------------------
MAB_2372|M.abscessus_ATCC_1997 PNVGKSTLVNRIIGRREAVVQDIPGVTRDRVSYPAEWLDRRFTVQDTGGW
Mflv_3493|M.gilvum_PYR-GCK EPDAKGLQQLVADQASVAMRTADAIIFVVDAVVGATTADEAAAKLLQRSG
Mvan_3274|M.vanbaalenii_PYR-1 EPDAKGLQQLVAEQASVAMRTADAIIFVVDSVVGATAADEAAAKLLQRSG
MMAR_2528|M.marinum_M EPDAKGLQQLVAEQASVAMRTADAVVLVVDATVGATTADEAAARILLRSG
MUL_3232|M.ulcerans_Agy99 EPDAKGLQQLVAEQASVAMRTADAVVLVVDATVGATTADEAAARILLRSG
Mb1740|M.bovis_AF2122/97 EPNAKGLQRLVAEQASVAMRTADAVILVVDAGVGATAADEAAARILLRSG
Rv1713|M.tuberculosis_H37Rv EPNAKGLQRLVAEQASVAMRTADAVILVVDAGVGATAADEAAARILLRSG
MLBr_01372|M.leprae_Br4923 EPDAKGLKRLVAEQASVAMRTADAVILVVDVGVGATDADEAAARILLRSG
MAV_3063|M.avium_104 EPDAKGLQQLVAEQASVAMRTADAVILVVDALVGATTADEAAARILLRSG
MSMEG_3738|M.smegmatis_MC2_155 EPDAKGLQQLVADQALVAMRTADAVILVVDAVVGATAADEAAAKLLRRSG
TH_0203|M.thermoresistible__bu --------------------------------------------------
MAB_2372|M.abscessus_ATCC_1997 EADATGLQQLVAEQARHAMATADLIILVVDATVGATSTDEEAAKLLRRSG
Mflv_3493|M.gilvum_PYR-GCK KPVFLAANKVDNERGEADAAALWSLGLGEPHPISAMHGRGVADLLDRVIE
Mvan_3274|M.vanbaalenii_PYR-1 KPVFLAANKVDNERGEADAAALWSLGLGEPHPISAMHGRGVADLLDAVVD
MMAR_2528|M.marinum_M KPVFLAANKVDSDKAEADAATLWSMGLGEPHAISAMHGRGVADLLDTVLK
MUL_3232|M.ulcerans_Agy99 KPVFLAANKVDSDKAEADAATLWSMGLGEPHAISAMHGRGVANLLDTVLK
Mb1740|M.bovis_AF2122/97 KPVFLAANKVDSEKGESDAAALWSLGLGEPHAISAMHGRGVADLLDGVLA
Rv1713|M.tuberculosis_H37Rv KPVFLAANKVDSEKGESDAAALWSLGLGEPHAISAMHGRGVADLLDGVLA
MLBr_01372|M.leprae_Br4923 KLVFLAANKVDGEKGESDASALWSLGLGEPHAISAMHGRGVADLLDKVLA
MAV_3063|M.avium_104 KPVFLAANKVDSDKAEADAAMLWSLGLGEPHPISAMHGRGVADLLDEVLA
MSMEG_3738|M.smegmatis_MC2_155 KPVFLAANKVDTERGEADAASLWSLGVGEPHPISAMHGRGVADLLDTVME
TH_0203|M.thermoresistible__bu --------------------------------------------------
MAB_2372|M.abscessus_ATCC_1997 KPVFLAANKVDSDRAEADAAVLWSLGLGEPMPISAMHGRGVADLLDRVVA
Mflv_3493|M.gilvum_PYR-GCK DLPTISEVNAGGGGGPRRVALVGKPNVGKSSLLNRLSGDERSVVHDVAGT
Mvan_3274|M.vanbaalenii_PYR-1 ALPTISEVAGAGGGGPRRVALVGKPNVGKSSLLNRLSGDERSVVHDVAGT
MMAR_2528|M.marinum_M KLPEVAESVSA-GGGPRRVALVGKPNVGKSSLLNKLAGDERSVVHDVAGT
MUL_3232|M.ulcerans_Agy99 KLPEVAESVSA-GGGPRRVALVGKPNVGKSSLLNKLAGDERSVVHDVAGT
Mb1740|M.bovis_AF2122/97 ALPEVGESASA-SGGPRRVALVGKPNVGKSSLLNKLAGDQRSVVHEAAGT
Rv1713|M.tuberculosis_H37Rv ALPEVGESASA-SGGPRRVALVGKPNVGKSSLLNKLAGDQRSVVHEAAGT
MLBr_01372|M.leprae_Br4923 ALPNVAESTSL-DGGLRRVALVGKPNVGKSSLLNKLAGDQRSVVHEAAGT
MAV_3063|M.avium_104 ALPEVSEVAPR-PGGPRRVALVGKPNVGKSSLLNKLAGDQRSVVHDVAGT
MSMEG_3738|M.smegmatis_MC2_155 KLPEVSEVATGGSGGPRRVALVGKPNVGKSSLLNRLAGDHRSVVHDMAGT
TH_0203|M.thermoresistible__bu --------------------------VGKSSLLNRLAGDQRAVVHDVAGT
MAB_2372|M.abscessus_ATCC_1997 DLPEISARGSG-EGGPRRVALVGKPNVGKSSLLNRLSGDQRAVVHDTAGT
********:*:**.*:***: ***
Mflv_3493|M.gilvum_PYR-GCK TVDPVDSLIEMDGKLWRFVDTAGLRRKVGQASGHEFYASVRTHGAIDSAE
Mvan_3274|M.vanbaalenii_PYR-1 TVDPVDSLIEMDGKLWRFVDTAGLRRKVGQASGHEFYASVRTHGAIDAAE
MMAR_2528|M.marinum_M TVDPVDSLIELGGKVWRFVDTAGLRRKVGQASGHEFYASVRTRGAIDAAE
MUL_3232|M.ulcerans_Agy99 TVDPVDSLIELGGKVWRFVDTAGLRRKVGQASGHEFYASVRTRGAIDAAE
Mb1740|M.bovis_AF2122/97 TVDPVDSLIELGGDVWRFVDTAGLRRKVGQASGHEFYASVRTHAAIDSAE
Rv1713|M.tuberculosis_H37Rv TVDPVDSLIELGGDVWRFVDTAGLRRKVGQASGHEFYASVRTHAAIDSAE
MLBr_01372|M.leprae_Br4923 TVDPVDSLIEMGGRVWRFVDTAGLRRKVGQASGHEFYASVRTHGAIDSAE
MAV_3063|M.avium_104 TVDPVDSLIELGDRVWRFVDTAGLRRKVGQASGHEFYASVRTHSAIDAAE
MSMEG_3738|M.smegmatis_MC2_155 TVDPVDSLIELGGKTWRFVDTAGLRRKVGQATGHEFYASVRTHGAIDAAE
TH_0203|M.thermoresistible__bu TVDPVDSLIELGGKTWRFIDTAGLRRKVGQASGHEYYASVRTHGAIDAAE
MAB_2372|M.abscessus_ATCC_1997 TVDPVDTLIELGGKTWRFVDTAGLRRKVGQASGHEYYASLRTHGAIEAAE
******:***:.. ***:************:***:***:**:.**::**
Mflv_3493|M.gilvum_PYR-GCK VAIVLIDASQPLTEQDQRVLSMVIEAGRALVLAFNKWDLVDEDRRYLLDR
Mvan_3274|M.vanbaalenii_PYR-1 VAIVLVDASQPLTEQDQRVLSMVVEAGRALVLAFNKWDLVDEDRRYLLDR
MMAR_2528|M.marinum_M VVVLLIDASQPLTEQDLRVLSMVIEAGRAVVLAYNKWDLVDEDRRDLLDR
MUL_3232|M.ulcerans_Agy99 VVVLLIDASQPLTEQDLRVLSMVIEAGRAVVLAYNKWDLVDEDRRDLLDR
Mb1740|M.bovis_AF2122/97 VAIVLIDASQPLTEQDLRVISMVIEAGRALVLAYNKWDLVDEDRRELLQR
Rv1713|M.tuberculosis_H37Rv VAIVLIDASQPLTEQDLRVISMVIEAGRALVLAYNKWDLVDEDRRELLQR
MLBr_01372|M.leprae_Br4923 VVIMLIDASEPLTGQDQRVLSMVIDAGRALVLAFNKWDLVDEDRCDLLER
MAV_3063|M.avium_104 VVIVLIDASAPLTEQDQRVLSMVIEAGRALVLAFNKWDLVDEDRRELLER
MSMEG_3738|M.smegmatis_MC2_155 VVIVLIDASQPLTEQDQRVLSMVIEAGRALVLAFNKWDLVDEDRRYLLSR
TH_0203|M.thermoresistible__bu VVIVLIDASQPVTEQDQRILSMVTDAGRALVLAFNKWDLVDEERRHLLER
MAB_2372|M.abscessus_ATCC_1997 VAIVLLDASQPITEQDQRVLSMVIESGRALVLAFNKWDLVDEDRRDLLER
*.::*:*** *:* ** *::*** ::***:***:********:* **.*
Mflv_3493|M.gilvum_PYR-GCK EIDLQLAQLQWAPRVNISAKTGRAVQKLVPALETSLASWDKRITTGQLNT
Mvan_3274|M.vanbaalenii_PYR-1 EIDLQLAQLQWAPRVNISAKTGRAVQKLVPALETALKSWDTRVSTGRLNT
MMAR_2528|M.marinum_M EIDRELVQIRWAQRVNISAKTGRAVQKLVPAMETALASWDTRIPTGPLNS
MUL_3232|M.ulcerans_Agy99 EIDRELVQIRWAQRVNISAKTGRAVQKLVPAMETALASWDTRIPTGPLNS
Mb1740|M.bovis_AF2122/97 EIDRELVQVRWAQRVNISAKTGRAVHKLVPAMEDALASWDTRIATGPLNT
Rv1713|M.tuberculosis_H37Rv EIDRELVQVRWAQRVNISAKTGRAVHKLVPAMEDALASWDTRIATGPLNT
MLBr_01372|M.leprae_Br4923 EIDRELVQVRWAQRVNISAKTGRAVQKLVPAMENSLASWDTRIATGPLNI
MAV_3063|M.avium_104 EIDRELVQLRWAPRVNISAKTGRAVAKLVPAMETALASWDTRIATGPLNS
MSMEG_3738|M.smegmatis_MC2_155 EIERELVQVQWAPRINISALTGRAVQKLVPALETSLTSWDTRISTGRLNT
TH_0203|M.thermoresistible__bu EIDMDLSHVHWAPRVNVSAKTGRAVQKLVPALERALASWDNRITTGQLNT
MAB_2372|M.abscessus_ATCC_1997 EVDLQLAQLNWAQRVNISAKSGRAVQKLVPALESALDSWDKRISTGQLNT
*:: :* ::.** *:*:** :**** *****:* :* ***.*:.** **
Mflv_3493|M.gilvum_PYR-GCK FLKEVVAATPPPVRGGKQPRILFATQATSRPPTFVLFTSGFLEAGYRRFL
Mvan_3274|M.vanbaalenii_PYR-1 FFKEIVAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEAGYRRFL
MMAR_2528|M.marinum_M WIKEVVAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEAGYRRFL
MUL_3232|M.ulcerans_Agy99 WIKEVVAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEAGYRRFL
Mb1740|M.bovis_AF2122/97 WLTEVTAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEAGYRRFL
Rv1713|M.tuberculosis_H37Rv WLTEVTAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEAGYRRFL
MLBr_01372|M.leprae_Br4923 WIKAVVAATPPPVRGGKQPRILFATQATARPPTFVLFTTGFLEACYRRFL
MAV_3063|M.avium_104 WLKEVVAATPPPVRGGKQPRILFATQAAARPPTFVLFTTGFLEAGYRRFL
MSMEG_3738|M.smegmatis_MC2_155 FLKEVVAATPPPVRGGKQPRILFATQAATRPPTFVLFTSGFLEAGYRRFL
TH_0203|M.thermoresistible__bu FLKEVLAATPPPVRGGRQPRVLFATQAAIRPPTFVLFTTGFLEAGYRRFL
MAB_2372|M.abscessus_ATCC_1997 WVKEVVAATPPPVRGGKQPRVLFATQATSRPPTFVLFTTGFLEAGYRRFL
:.. : **********:***:******: *********:***** *****
Mflv_3493|M.gilvum_PYR-GCK ERRLRETFGFEGSPIRINVRVREKRGPKSRR--
Mvan_3274|M.vanbaalenii_PYR-1 ERRLRETFGFEGSPIRINVRVREKRGPKARR--
MMAR_2528|M.marinum_M ERRLRETFGFEGSPIRINVRVREKRGAKRPGRR
MUL_3232|M.ulcerans_Agy99 ERRLRETFGFEGSPIRINVRVREKRGAKRPGRR
Mb1740|M.bovis_AF2122/97 ERRLRETFGFDGSPIRVNVRVREKRAGKRR---
Rv1713|M.tuberculosis_H37Rv ERRLRETFGFDGSPIRVNVRVREKRAGKRR---
MLBr_01372|M.leprae_Br4923 ERRLRETFGFEGSPIRINVRVREKRGLKRR---
MAV_3063|M.avium_104 ERRLREAFGFEGTPIRINVRVREKRGARRR---
MSMEG_3738|M.smegmatis_MC2_155 ERKLRETFGFEGSPIRVNVRVREKR-AKR----
TH_0203|M.thermoresistible__bu ERRLREEFGFEGSPLRINVRVREKRAKK-----
MAB_2372|M.abscessus_ATCC_1997 ERRLRESFGFDGTPIRINVRVREKRSAKR----
**:*** ***:*:*:*:******** :