For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNGTESSAAEPQNGYPDGFRVRLTNFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTKAIGAQLDLE ETTAFLVVAATLLDLKAARLLPAGRMDDEEDLALLEVRDLLFARLLQYRAFKHVAEMFAELEATALRSYP RGVSLEDRYATLLPEVMLGVDAERFAQIAAVALTPRPVPTVAIGHLHEQTVSVPEQAKHLLTMLESRGTG QWASFSELVADCTAPMEIVGRFLALLELYRSRAVAFDQSEPLGVLQVSWTGERPTSEALVEVRDQ
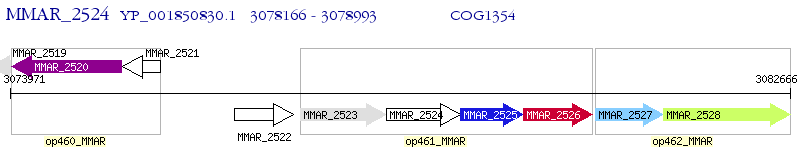
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2524 | - | - | 100% (275) | hypothetical protein MMAR_2524 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1736 | - | 1e-125 | 84.59% (266) | hypothetical protein Mb1736 |
| M. gilvum PYR-GCK | Mflv_3497 | - | 1e-113 | 79.53% (254) | chromosome segregation and condensation protein ScpA |
| M. tuberculosis H37Rv | Rv1709 | - | 1e-125 | 84.59% (266) | hypothetical protein Rv1709 |
| M. leprae Br4923 | MLBr_01368 | - | 1e-115 | 78.54% (261) | hypothetical protein MLBr_01368 |
| M. abscessus ATCC 19977 | MAB_2368 | - | 1e-107 | 70.37% (270) | hypothetical protein MAB_2368 |
| M. avium 104 | MAV_3067 | scpA | 1e-126 | 82.55% (275) | segregation and condensation protein A |
| M. smegmatis MC2 155 | MSMEG_3742 | scpA | 1e-116 | 82.94% (252) | segregation and condensation protein A |
| M. thermoresistible (build 8) | TH_0198 | - | 1e-117 | 78.91% (275) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3236 | - | 1e-153 | 100.00% (275) | hypothetical protein MUL_3236 |
| M. vanbaalenii PYR-1 | Mvan_3278 | - | 1e-117 | 79.78% (267) | chromosome segregation and condensation protein ScpA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3497|M.gilvum_PYR-GCK MSEPVS------------------------PDEPATESTPAG----FQVR
Mvan_3278|M.vanbaalenii_PYR-1 MSETT-------------------------PEEPTDSSTQVG----FQVR
MSMEG_3742|M.smegmatis_MC2_155 MNDDVREPDQDPASVAAPEDNVSSDAASDTAEDTAEDEANKG----FKVR
TH_0198|M.thermoresistible__bu VSGQPD------------------------SDTGHHDEAATG----FQVR
MMAR_2524|M.marinum_M MNGTES------------------------SAAE----PQNGYPDGFRVR
MUL_3236|M.ulcerans_Agy99 MNGTES------------------------SAAE----PQNGYPDGFRVR
Mb1736|M.bovis_AF2122/97 MNGLQN------------------------SLANGGTAPENGYSAGFRVR
Rv1709|M.tuberculosis_H37Rv MNGLQN------------------------SLANGGTAPENGYSAGFRVR
MAV_3067|M.avium_104 MNATAN------------------------GEAQ----PQTG----FQVR
MLBr_01368|M.leprae_Br4923 MN----------------------------SEAP----HQNG----FQVR
MAB_2368|M.abscessus_ATCC_1997 MTVGLDQ-----------EPDVVETAAEPAAPDAATAAPETTDAHGFRIR
:. *::*
Mflv_3497|M.gilvum_PYR-GCK LSNFEGPFDLLLQLIFGHRLDVTEVALHQVTDEFIAYTKQIGPQLGLDET
Mvan_3278|M.vanbaalenii_PYR-1 LSNFEGPFDLLLQLIFAHRLDVTEVALHQVTDEFIAYTKEIGPQLGLDET
MSMEG_3742|M.smegmatis_MC2_155 LTNFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTKEIGPQLGLDET
TH_0198|M.thermoresistible__bu LANFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTRQIGRELSLEET
MMAR_2524|M.marinum_M LTNFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTKAIGAQLDLEET
MUL_3236|M.ulcerans_Agy99 LTNFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTKAIGAQLDLEET
Mb1736|M.bovis_AF2122/97 LTNFEGPFDLLLQLIFAHQLDVTEVALHQVTDDFIAYTKAIGARLELEET
Rv1709|M.tuberculosis_H37Rv LTNFEGPFDLLLQLIFAHQLDVTEVALHQVTDDFIAYTKAIGARLELEET
MAV_3067|M.avium_104 LTNFEGPFDLLLQLIFAHRLDVTEVALHQVTDDFIAYTREIGPKLELEET
MLBr_01368|M.leprae_Br4923 LTNFEGPFDLLLHLICAHRLDVTEVALHQVTDDFIAYIRRLGPQLGLEET
MAB_2368|M.abscessus_ATCC_1997 LTNFEGPFDLLLQLISQHRLDVTEVALHQVTDEFIAYTKSLGDTYSLDEV
*:**********:** *:*************:**** : :* *:*.
Mflv_3497|M.gilvum_PYR-GCK TSFLVVAATLLDLKAARLLPAGEIHDEEDLALLEVRDLLFARLLQYRAFK
Mvan_3278|M.vanbaalenii_PYR-1 TSFLVVAATLLDLKAARLLPAGEIHDEDDLALLEVRDLLFARLLQYRAFK
MSMEG_3742|M.smegmatis_MC2_155 TAFLVIAATLLDLKAARLLPSGEVHDEEDLALLEVRDLLFARLLQYRAFK
TH_0198|M.thermoresistible__bu TAFLVVAATLLDLKAARLLPDGSVHDEEDLALLEVRDLLFARLLQYRAFK
MMAR_2524|M.marinum_M TAFLVVAATLLDLKAARLLPAGRMDDEEDLALLEVRDLLFARLLQYRAFK
MUL_3236|M.ulcerans_Agy99 TAFLVVAATLLDLKAARLLPAGRMDDEEDLALLEVRDLLFARLLQYRAFK
Mb1736|M.bovis_AF2122/97 TAFLVIAATLLDLKAARLLPAGQVDDEEDLALLEVRDLLFARLLQYRAFK
Rv1709|M.tuberculosis_H37Rv TAFLVIAATLLDLKAARLLPAGQVDDEEDLALLEVRDLLFARLLQYRAFK
MAV_3067|M.avium_104 TAFLVVAATLLDLKAARLLPAGQVDDDEDLALLEVRDLLFARLLQYRAFK
MLBr_01368|M.leprae_Br4923 TAFLVIAATLIDLKAARLLPAGRVEDEEDLALLEGRDLLFARLLQYRAFK
MAB_2368|M.abscessus_ATCC_1997 TAFLVVAATLLDLKAARLLPSGQVDNDDDLALLEIRDLLFARLLQYRAFK
*:***:****:********* * :.:::****** ***************
Mflv_3497|M.gilvum_PYR-GCK HVAEMFAELEAAAMRSYPRAVSLEDRYSDLLPEVMLGVDATRFAEIAAAA
Mvan_3278|M.vanbaalenii_PYR-1 HVAEMFAELEAAALRSYPRAVSLEDTYSDLLPEVMLGVDAARFAEIAAAA
MSMEG_3742|M.smegmatis_MC2_155 HVAQMFAELEAAALRSYPRAVSLEDRYSDLLPEVMLGVDAEKFATIAAAA
TH_0198|M.thermoresistible__bu HVAEMFAELEAAALRSYPRSVSLEERYTRLLPEVMLGVDAERFAEIAAAA
MMAR_2524|M.marinum_M HVAEMFAELEATALRSYPRGVSLEDRYATLLPEVMLGVDAERFAQIAAVA
MUL_3236|M.ulcerans_Agy99 HVAEMFAELEATALRSYPRGVSLEDRYATLLPEVMLGVDAERFAQIAAVA
Mb1736|M.bovis_AF2122/97 HVAEMFAELEATALRSYPRAVSLEDGFVGLLPEVMLGVDAHRFAEIAAIA
Rv1709|M.tuberculosis_H37Rv HVAEMFAELEATALRSYPRAVSLEDGFVGLLPEVMLGVDAHRFAEIAAIA
MAV_3067|M.avium_104 HVAEMFAELEASALRSYPRAVSLEDRFTQLLPEVMLGVDAERFAQIAAVA
MLBr_01368|M.leprae_Br4923 RVAQIFAELEATALRSYPHAVFLEERFASLLPEVMIGVDADRFAEIAAFT
MAB_2368|M.abscessus_ATCC_1997 HVAEMFAELEAAALRSYPRAVSLEERFSDLLPPVHLGLDGDQFAQLAAGA
:**::******:*:****:.* **: : *** * :*:*. :** :** :
Mflv_3497|M.gilvum_PYR-GCK FTPRPVPSVGLDHLHQVSVSVPQQAANLIALLERRGIGQWASFTDLVSDC
Mvan_3278|M.vanbaalenii_PYR-1 FTPRPVPTVGTEHLHQVAVSVPEQAANLMTLLEQRGIGHWASFSELVADC
MSMEG_3742|M.smegmatis_MC2_155 FTPRPVPTVATDHVHAPKVSVPEQAYRILALLEQRGVGEWASFSELVAEC
TH_0198|M.thermoresistible__bu LTPRPVPTVATEHLHAPKVSVPEQAKRLLSLLERRGPGQWASFGELVADC
MMAR_2524|M.marinum_M LTPRPVPTVAIGHLHEQTVSVPEQAKHLLTMLESRGTGQWASFSELVADC
MUL_3236|M.ulcerans_Agy99 LTPRPVPTVAIGHLHEQTVSVPEQAKHLLTMLESRGTGQWASFSELVADC
Mb1736|M.bovis_AF2122/97 LTPRPAPTVATEHLHELMVSVPEQAEHLLAMLKARGSGQWASFSELVADC
Rv1709|M.tuberculosis_H37Rv LTPRPAPTVATEHLHELMVSVPEQAEHLLAMLKARGSGQWASFSELVADC
MAV_3067|M.avium_104 FSPRPVPTVSVGHLHEVKVSVPEQARKLLAILEARGSGQWATFSELVADC
MLBr_01368|M.leprae_Br4923 FTPRPVPIVATGHLHELKISVPEQAKRLLAMLEARGNGQWTSFSELVADC
MAB_2368|M.abscessus_ATCC_1997 FTPRPVPTVGLSHLHIPRVSVPEHAKRMVELLAERGAGQWMTFTELVAEC
::***.* *. *:* :***::* .:: :* ** *.* :* :**::*
Mflv_3497|M.gilvum_PYR-GCK QVPIEIVGRFLALLELYRARAVTFSQPEALGVLQISWTGERPDSQQLATA
Mvan_3278|M.vanbaalenii_PYR-1 EAPMEIVGRFLALLELYRARAVAFEQPEPLGVLQISWTGERPNSQQLATA
MSMEG_3742|M.smegmatis_MC2_155 TDGLQIVGRFLALLELFRARAVAFEQSEPLGVLQVSWTGDRPTSEHLAAA
TH_0198|M.thermoresistible__bu QTGLEIVGRFLALLELYRARTVAFEQSEPLGVLQVSWTGERPTDEHLATA
MMAR_2524|M.marinum_M TAPMEIVGRFLALLELYRSRAVAFDQSEPLGVLQVSWTGERPTSEALVEV
MUL_3236|M.ulcerans_Agy99 TAPMEIVGRFLALLELYRSRAVAFDQSEPLGVLQVSWTGERPTSEALVEV
Mb1736|M.bovis_AF2122/97 TAPIEIVGRFLALLELYRTRAVAFEQSEPLGALQVSWTGDDAERSDEKER
Rv1709|M.tuberculosis_H37Rv TAPIEIVGRFLALLELYRTRAVAFEQSEPLGALQVSWTGDDAERSDEKER
MAV_3067|M.avium_104 EGSMEVVGRFLALLELYRSRAVAFEQSEPLGVLQISWTGERPVGETLVEV
MLBr_01368|M.leprae_Br4923 REPIEVVGCFLALLELYRTRAIAFEQAEPLGVLQVSWAGERLASEALAEV
MAB_2368|M.abscessus_ATCC_1997 TAPIEIVARFLGVLELYRSKAVLFEQSEPLGPLQVSWTGERPAQDDLEKA
:::*. **.:***:*:::: *.*.*.** **:**:*: .
Mflv_3497|M.gilvum_PYR-GCK DAE---
Mvan_3278|M.vanbaalenii_PYR-1 DAE---
MSMEG_3742|M.smegmatis_MC2_155 DAEE--
TH_0198|M.thermoresistible__bu ESEDDD
MMAR_2524|M.marinum_M RDQ---
MUL_3236|M.ulcerans_Agy99 RDQ---
Mb1736|M.bovis_AF2122/97 RL----
Rv1709|M.tuberculosis_H37Rv RL----
MAV_3067|M.avium_104 RDEL--
MLBr_01368|M.leprae_Br4923 RDES--
MAB_2368|M.abscessus_ATCC_1997 VDYT--