For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEDMEVVRQRALDCGAVEAVVVDARDEFAE GYCLPTILNNALYMDRYPLVSAISRPLIVKHLVEAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLE VLAPVRDYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNAPTKDVYSYTEDPTV NWNTPDEVIVGFERGMPVSIDGNAVTMLQAIEELNRRAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVL ITAHAELEHVTLERELARFKRHTDQRWAELVYDGLWYSPLKTALESFVAKTQEHVSGEIRLVLHGGHIAV NGRRSAESLYDFNLATYDEGDTFDQSAAKGFVYVHGLSSKLSARRDLQ
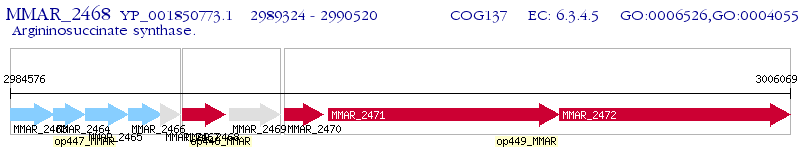
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2468 | argG | - | 100% (398) | argininosuccinate synthase ArgG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1686 | argG | 0.0 | 93.47% (398) | argininosuccinate synthase |
| M. gilvum PYR-GCK | Mflv_3524 | - | 0.0 | 91.94% (397) | argininosuccinate synthase |
| M. tuberculosis H37Rv | Rv1658 | argG | 0.0 | 93.47% (398) | argininosuccinate synthase |
| M. leprae Br4923 | MLBr_01412 | argG | 0.0 | 90.95% (398) | argininosuccinate synthase |
| M. abscessus ATCC 19977 | MAB_2342 | - | 0.0 | 88.64% (396) | argininosuccinate synthase |
| M. avium 104 | MAV_3112 | argG | 0.0 | 93.94% (396) | argininosuccinate synthase |
| M. smegmatis MC2 155 | MSMEG_3770 | argG | 0.0 | 92.19% (397) | argininosuccinate synthase |
| M. thermoresistible (build 8) | TH_1598 | argG | 0.0 | 91.21% (398) | Probable Argininosuccinate synthase argG |
| M. ulcerans Agy99 | MUL_1650 | argG | 0.0 | 99.50% (398) | argininosuccinate synthase |
| M. vanbaalenii PYR-1 | Mvan_3308 | - | 0.0 | 91.69% (397) | argininosuccinate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3524|M.gilvum_PYR-GCK ----MSERVILAYSGGLDTSVAISWIGKETGKEVVAVAIDLGQGGEDMEV
Mvan_3308|M.vanbaalenii_PYR-1 ----MSERVILAYSGGLDTSVAISWIGKETGSEVVAVAIDLGQGGEDMEV
TH_1598|M.thermoresistible__bu ----MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEDMEV
MAB_2342|M.abscessus_ATCC_1997 MPVEHSDRVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEDMEV
MSMEG_3770|M.smegmatis_MC2_155 ----MSERVILAYSGGLDTSVAISWIGKETGKEVVAVAIDLGQGGEDMEV
MMAR_2468|M.marinum_M ----MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEDMEV
MUL_1650|M.ulcerans_Agy99 ----MSERVILAYSGGLDTSVAISWIGEETGREVVAVAIDLGQGGEDMEV
Mb1686|M.bovis_AF2122/97 ----MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEHMDV
Rv1658|M.tuberculosis_H37Rv ----MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEHMDV
MLBr_01412|M.leprae_Br4923 ---MMPERIILAYSGGLDTSVAINWIGKETSHEVVAVVIDLGQGGEDMEV
MAV_3112|M.avium_104 ----MSERVILAYSGGLDTSVAISWIGKETGREVVAVAIDLGQGGEDMEV
.:*:**************.***:**. *****.********.*:*
Mflv_3524|M.gilvum_PYR-GCK VRQRAIDCGAVEAVVVDARDEFAEQYCLPAIQSNALYMDRYPLVSALSRP
Mvan_3308|M.vanbaalenii_PYR-1 VRQRALDCGAVEAVVVDARDEFAEQYCLPAIKSNALYMDRYPLVSALSRP
TH_1598|M.thermoresistible__bu VRQRALDCGAVESIVIDARDEFANDYCVPAIQSNALYMDRYPLVSALSRP
MAB_2342|M.abscessus_ATCC_1997 VRQRALDCGAVEAVVVDARDEFAEEYCLPTIQSNALYMDRYPLVSAISRP
MSMEG_3770|M.smegmatis_MC2_155 VRQRALDCGAVEAVVVDARDEFAEEYCLPTIQSNALYMDRYPLVSAISRP
MMAR_2468|M.marinum_M VRQRALDCGAVEAVVVDARDEFAEGYCLPTILNNALYMDRYPLVSAISRP
MUL_1650|M.ulcerans_Agy99 VCQRALDCGAVEAVVVDARDEFAEGYCLPTILNNALYMDRYPLVSAISRP
Mb1686|M.bovis_AF2122/97 IRQRALDCGAVEAVVVDARDEFAEGYCLPTVLNNALYMDRYPLVSAISRP
Rv1658|M.tuberculosis_H37Rv IRQRALDCGAVEAVVVDARDEFAEGYCLPTVLNNALYMDRYPLVSAISRP
MLBr_01412|M.leprae_Br4923 VRQRALDCGAVEAIVVDARDEFAEGYCLPTVLNNALYMDRYPLVSAISRP
MAV_3112|M.avium_104 IRQRALDCGAVEAVVVDARDEFAEGYCLPTIRNNALYMDRYPLVSAISRP
: ***:******::*:*******: **:*:: .*************:***
Mflv_3524|M.gilvum_PYR-GCK LIVKHLVDAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLKVLAPVR
Mvan_3308|M.vanbaalenii_PYR-1 LIVKHLVDAAREHGGGVVAHGCTGKGNDQVRFEVGFASLAPDLKVLAPVR
TH_1598|M.thermoresistible__bu LIVKHLVKAAREHGGTIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
MAB_2342|M.abscessus_ATCC_1997 LIVKHLVAAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
MSMEG_3770|M.smegmatis_MC2_155 LIVKHLVKAAREHGGGVVAHGCTGKGNDQVRFEVGFASLAPDLQVLAPVR
MMAR_2468|M.marinum_M LIVKHLVEAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
MUL_1650|M.ulcerans_Agy99 LIVKHLVEAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
Mb1686|M.bovis_AF2122/97 LIVKHLVAAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
Rv1658|M.tuberculosis_H37Rv LIVKHLVAAAREHGGGIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
MLBr_01412|M.leprae_Br4923 LIVKHLVAAARAHGGSIVAHGCTGKGNDQVRFEVGFASLAPDLEILAPVR
MAV_3112|M.avium_104 LIVKHLVAAAREHGGSIVAHGCTGKGNDQVRFEVGFASLAPDLEVLAPVR
******* *** *** :**************************::*****
Mflv_3524|M.gilvum_PYR-GCK DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
Mvan_3308|M.vanbaalenii_PYR-1 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
TH_1598|M.thermoresistible__bu DYAWTREKAIAFAEENNIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
MAB_2342|M.abscessus_ATCC_1997 DYAWTREKAIAFAAENEIPINVTKKSPFSIDQNVWGRAVETGFLEDLWNA
MSMEG_3770|M.smegmatis_MC2_155 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
MMAR_2468|M.marinum_M DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
MUL_1650|M.ulcerans_Agy99 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
Mb1686|M.bovis_AF2122/97 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
Rv1658|M.tuberculosis_H37Rv DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
MLBr_01412|M.leprae_Br4923 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWHA
MAV_3112|M.avium_104 DYAWTREKAIAFAEENAIPINVTKRSPFSIDQNVWGRAVETGFLEHLWNA
************* ** *******:********************.**:*
Mflv_3524|M.gilvum_PYR-GCK PTKDVYDYTEDPTVNWSSPDEVVVGFEKGVPVSIDGNPVTVLQAIEQLNE
Mvan_3308|M.vanbaalenii_PYR-1 PTKDVYDYTEDPTLNWSSPDEVIIGFDKGVPVSIDGKPVTVLQAIEQLNA
TH_1598|M.thermoresistible__bu PTKDVYSYTEDPTVNWSTPDEVIVGFEQGVPVSIDGRSVTPLQAIEELNR
MAB_2342|M.abscessus_ATCC_1997 PTKDVYDYTEDPTLNWGAPDELIVTFDKGVPTAIDGHPVSVLEAIVELNR
MSMEG_3770|M.smegmatis_MC2_155 PTKDVYDYTEDPTVHWNTPDEVVIGFDKGVPVSIDGRPVSVLQAIEELNR
MMAR_2468|M.marinum_M PTKDVYSYTEDPTVNWNTPDEVIVGFERGMPVSIDGNAVTMLQAIEELNR
MUL_1650|M.ulcerans_Agy99 PTKDVYSYTEDPTVNWNTPDEVIVGFERGMPVSIDGNAVTMLQAIEELNR
Mb1686|M.bovis_AF2122/97 PTKDIYAYTEDPTINWGVPDEVIVGFERGVPVSVDGKPVSMLAAIEELNR
Rv1658|M.tuberculosis_H37Rv PTKDIYAYTEDPTINWGVPDEVIVGFERGVPVSVDGKPVSMLAAIEELNR
MLBr_01412|M.leprae_Br4923 PTKEVYSYTDDPTINWNTPDEVIVGFEHGVPVSIDGSPVSMLGAIEALNR
MAV_3112|M.avium_104 PTKDVYAYTEDPTLNWSTPDEVIVGFERGVPVSIDGKPVSVLGAIEELNA
***::* **:***::*. ***::: *::*:*.::** .*: * ** **
Mflv_3524|M.gilvum_PYR-GCK RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
Mvan_3308|M.vanbaalenii_PYR-1 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
TH_1598|M.thermoresistible__bu RGGEQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
MAB_2342|M.abscessus_ATCC_1997 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHEELEHVTLEREL
MSMEG_3770|M.smegmatis_MC2_155 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
MMAR_2468|M.marinum_M RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHAELEHVTLEREL
MUL_1650|M.ulcerans_Agy99 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHAELEHVTLEREL
Mb1686|M.bovis_AF2122/97 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
Rv1658|M.tuberculosis_H37Rv RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
MLBr_01412|M.leprae_Br4923 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHAELEHVTLEREL
MAV_3112|M.avium_104 RAGAQGVGRLDVVEDRLVGIKSREIYEAPGAMVLITAHTELEHVTLEREL
*.* ********************************** ***********
Mflv_3524|M.gilvum_PYR-GCK GRYKRLTDQKWGELVYDGLWFSPLKSALESFVANTQEHVSGEIRLVLHGG
Mvan_3308|M.vanbaalenii_PYR-1 GRFKRTTDQKWGELVYDGLWFSPLKTALESFVANTQEHVTGEIRLVLHGG
TH_1598|M.thermoresistible__bu GRFKRITDQKWGELVYDGLWFSPLKTALESFVAKTQEHVTGEIRMVLHGG
MAB_2342|M.abscessus_ATCC_1997 GRYKRRTDQKWAELVYDGLWYSPLKRSLEAFVADTQQHVSGDIRLVLHGG
MSMEG_3770|M.smegmatis_MC2_155 GRFKRHTDQKWGELVYDGLWYSPLKRALESFVAHTQEHVSGEIRLVLHGG
MMAR_2468|M.marinum_M ARFKRHTDQRWAELVYDGLWYSPLKTALESFVAKTQEHVSGEIRLVLHGG
MUL_1650|M.ulcerans_Agy99 ARFKRHTDQRWAELVYDGLWYSPLKTALESFVAKTQEHVSGEIRLVLHGG
Mb1686|M.bovis_AF2122/97 GRFKRQTDQRWAELVYDGLWYSPLKAALEAFVAKTQEHVSGEVRLVLHGG
Rv1658|M.tuberculosis_H37Rv GRFKRQTDQRWAELVYDGLWYSPLKAALEAFVAKTQEHVSGEVRLVLHGG
MLBr_01412|M.leprae_Br4923 GRFKRQTDRRWAELVYDGLWYSPLKTALESFVAATQQHVTGEVRMVLHGG
MAV_3112|M.avium_104 GRFKRHTDQRWAELVYDGLWYSPLKAALESFVDKTQEHVTGEIRMVLHGG
.*:** **::*.********:**** :**:** **:**:*::*:*****
Mflv_3524|M.gilvum_PYR-GCK HIAVNGRRSSESLYDFNLATYDEGDTFDQSSAKGFVHVHGLSSSLSARRD
Mvan_3308|M.vanbaalenii_PYR-1 HIAVNGRRSQESLYDFNLATYDEGDTFDQSAAKGFVHVHGLSSSLSARRD
TH_1598|M.thermoresistible__bu HIAVNGRRSPKSLYDFNLATYDEGDTFDQSAAKGFVQIHGLSSSISARRD
MAB_2342|M.abscessus_ATCC_1997 NIAVNGRRSPESLYDFNLATYDEGDSFDQSSAKGFVHIHGLSSKISAQRD
MSMEG_3770|M.smegmatis_MC2_155 HISVNGRRSAESLYDFNLATYDEGDSFDQSAAKGFVHLHGLSSRISAKRD
MMAR_2468|M.marinum_M HIAVNGRRSAESLYDFNLATYDEGDTFDQSAAKGFVYVHGLSSKLSARRD
MUL_1650|M.ulcerans_Agy99 HIAVNGRRSAESLYDFNLATYDEGDTFDQSAAKGFVYVHGLSSKLSARRD
Mb1686|M.bovis_AF2122/97 HIAVNGRRSAESLYDFNLATYDEGDSFDQSAARGFVYVHGLSSKLAARRD
Rv1658|M.tuberculosis_H37Rv HIAVNGRRSAESLYDFNLATYDEGDSFDQSAARGFVYVHGLSSKLAARRD
MLBr_01412|M.leprae_Br4923 HIAVNGRRSAESLYDFNLATYDEGDTFDQSAARGFVYVYGLPSKLAARRD
MAV_3112|M.avium_104 HIAVNGRRSAESLYDFNLATYDEGDSFDQSAAKGFVYVHGLSSKIASRRD
:*:****** :**************:****:*:*** ::**.* ::::**
Mflv_3524|M.gilvum_PYR-GCK LAGK
Mvan_3308|M.vanbaalenii_PYR-1 LAGK
TH_1598|M.thermoresistible__bu LQGQ
MAB_2342|M.abscessus_ATCC_1997 LSGK
MSMEG_3770|M.smegmatis_MC2_155 LGI-
MMAR_2468|M.marinum_M LQ--
MUL_1650|M.ulcerans_Agy99 LQ--
Mb1686|M.bovis_AF2122/97 LR--
Rv1658|M.tuberculosis_H37Rv LR--
MLBr_01412|M.leprae_Br4923 LR--
MAV_3112|M.avium_104 QR--