For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLAARLVTLVEQASAILDAAVPRFLGGHRADSAVPKKGNDFATEVDLAIERQVVAALEAATGIGVHGEE FGGTDVDSPWVWVLDPVDGTFNYAAGSPMAAILLGLLHEGEPVAGLTWLPFIGERYTAVAGGPLLRNGLP QASLAPAKLSESLMGVGTFSADSRGRFPGRYRLAVLENLSRVSSRLRIHGSTGIDLVYVADGILGGAISF GGQVWDHAAGVAQVRAAGGTVTTLTGDPWTPTSRSVLAAAPGVHEEILEIVRKTGDPEDY
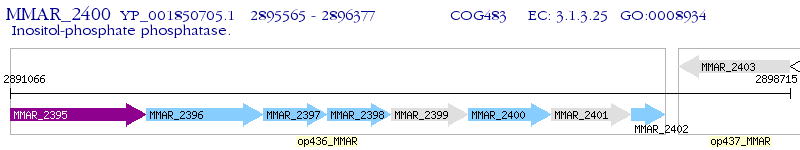
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2400 | impA | - | 100% (270) | inositol-monophosphatase ImpA |
| M. marinum M | MMAR_1511 | - | 9e-20 | 33.48% (230) | monophosphatase |
| M. marinum M | MMAR_2013 | suhB | 1e-16 | 31.25% (272) | extragenic suppressor protein SuhB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1630 | impA | 1e-117 | 77.41% (270) | inositol-monophosphatase |
| M. gilvum PYR-GCK | Mflv_3608 | - | 1e-103 | 67.29% (266) | inositol-phosphate phosphatase |
| M. tuberculosis H37Rv | Rv1604 | impA | 1e-118 | 78.15% (270) | inositol-monophosphatase |
| M. leprae Br4923 | MLBr_00662 | - | 1e-18 | 33.77% (231) | putative monophosphatase |
| M. abscessus ATCC 19977 | MAB_2665c | - | 1e-100 | 65.19% (270) | inositol monophosphatase family protein |
| M. avium 104 | MAV_3182 | - | 1e-112 | 74.44% (270) | inositol monophosphatase family protein |
| M. smegmatis MC2 155 | MSMEG_3210 | - | 1e-109 | 72.45% (265) | inositol monophosphate phosphatase |
| M. thermoresistible (build 8) | TH_1158 | impA | 1e-105 | 69.55% (266) | PROBABLE INOSITOL-MONOPHOSPHATASE IMPA (IMP) |
| M. ulcerans Agy99 | MUL_1577 | impA | 1e-153 | 99.26% (270) | inositol-monophosphatase ImpA |
| M. vanbaalenii PYR-1 | Mvan_2809 | - | 1e-105 | 69.06% (265) | inositol-phosphate phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2400|M.marinum_M ----MDLA-ARLVTLVEQASAILDAAVPRFLGGHRADSAVPKKGNDFATE
MUL_1577|M.ulcerans_Agy99 ----MDLA-ARLVTLVEQASAILDAAVPRFLGGHRADSAVPKKGNDFATE
Mb1630|M.bovis_AF2122/97 ----MHLD-SLVAPLVEQASAILDAATALFLVGHRADSAVRKKGNDFATE
Rv1604|M.tuberculosis_H37Rv ----MHLD-SLVAPLVEQASAILDAATALFLVGHRADSAVRKKGNDFATE
MAV_3182|M.avium_104 ----MDLD-ALVA----RASAILDDASKPFLAGHRADSAVRKKGNDFATD
Mflv_3608|M.gilvum_PYR-GCK ----MTLDRSQLVDLLTTAAGVLDGASAQFIDGHRADSAVAKKGNDFATE
Mvan_2809|M.vanbaalenii_PYR-1 ----MSPEPQQLAELLDTAAGVLEAASAQFIAGHRADSAVAKKGNDFATE
MSMEG_3210|M.smegmatis_MC2_155 MTVVGELDPQKLTALVATAAEILDAASVPFVAGHRADSAVRKQGNDFATE
TH_1158|M.thermoresistible__bu ----MALDDADLEALVSQAAKILDAASVRFVEGHRADSAVPKQGNDFATE
MAB_2665c|M.abscessus_ATCC_199 ----MTTT-AELEALVREASVILDAAAQRFVAGHGAEGVVFKGGKDFATE
MLBr_00662|M.leprae_Br4923 ----------MLALTLADRADALTSARFGALN-LRVDTKPDLTP---VTD
: : * * : .: .*:
MMAR_2400|M.marinum_M VDLAIERQVVAALEAATG-IGVHGEEFGGTDVDSPWVWVLDPVDGTFNYA
MUL_1577|M.ulcerans_Agy99 VDLAIERQVVAALVAATG-IGVHGEEFGGTDVDSPWVWVLDPVDGTFNYA
Mb1630|M.bovis_AF2122/97 VDLAIERQVVAALVAATG-IEVHGEEFGGPAVDSRWVWVLDPIDGTINHA
Rv1604|M.tuberculosis_H37Rv VDLAIERQVVAALVAATG-IEVHGEEFGGPAVDSRWVWVLDPIDGTINYA
MAV_3182|M.avium_104 VDLAIERQVVAALVDATG-IGVHGEEFGGSAVDSEWVWVLDPVDGTFNYA
Mflv_3608|M.gilvum_PYR-GCK VDLAIERRVVAELNRLTG-IGVHGEEFGGESLESEQVWVLDPIDGTFNYA
Mvan_2809|M.vanbaalenii_PYR-1 IDLAIERRVVAELTRLTG-IGVHGEEFGGESVEAEYVWLLDPVDGTFNYA
MSMEG_3210|M.smegmatis_MC2_155 VDLAIERQVVRALTEATG-IGVHGEEFGGEPIDSPLVWVLDPIDGTFNYA
TH_1158|M.thermoresistible__bu VDLAIERQVVEALTGATG-IGVHGEEFGGADVTSPLVWVLDPIDGTFNYA
MAB_2665c|M.abscessus_ATCC_199 EDLAIERMVVQALTERTG-IGVHGEEFGGPAVDSGLVWVVDPIDGTVNYA
MLBr_00662|M.leprae_Br4923 ADRAVEADVRAVLGRERPKDGILGEEYGGTITFSGQQWIVDPIDGTKNFV
* *:* * * : ***:** : *::**:*** *..
MMAR_2400|M.marinum_M AGSPMAAILLGLLHEGEPVAGLTWLPFIGERYTAVAG-GPLLRNGLPQAS
MUL_1577|M.ulcerans_Agy99 AGSPMAAILLGLMHEGEPVAGLTWLPFIGERYTAVAG-GPLLRNGLPQAS
Mb1630|M.bovis_AF2122/97 AGSPLAAILLGLLHDGVPVAGLTWMPFTDQRYTAVAG-GPLIKNGVPQPP
Rv1604|M.tuberculosis_H37Rv AGSPLAAILLGLLHDGVPVAGLTWMPFTDPRYTAVAG-GPLIKNGVPQPP
MAV_3182|M.avium_104 AGSPMAGILLALLHHGDPVAGLTWLPFLDQRYTAVTG-GPLRKNEIPRPP
Mflv_3608|M.gilvum_PYR-GCK AGLPTAAILLGLLIDGEPVLGLTWLPFMDLRYTAILG-EQVRCNGAPLPG
Mvan_2809|M.vanbaalenii_PYR-1 AGSPMAAILLSLLWEGKPLLGLTWLPFMGQRYTSMVG-APVRCNGEELPR
MSMEG_3210|M.smegmatis_MC2_155 AGSPMAAILLGLLADGEPVAGLTWLPFTGEKYSALVG-GPLYSDGKPCPP
TH_1158|M.thermoresistible__bu AGLPLAAILLALLQDGEPVAGLTWMPFTDQRFTAIAG-KPLYLNGVEQPR
MAB_2665c|M.abscessus_ATCC_199 AGSPMAAILLGLMSDGVPVAGLTWLPFMGEKYAAVTG-GPLFRNGVAMPP
MLBr_00662|M.leprae_Br4923 RGVPVWASLIALLEDGVPSIGVVSAPALQRRWWAARGQGAFVAVDGVPRR
* * . *:.*: .* * *:. * :: : * .
MMAR_2400|M.marinum_M LAPAKLSESLMGVGTFSADSRGRFPGRYRLAVLENLSRVSSRLRIHGSTG
MUL_1577|M.ulcerans_Agy99 LAPAKLSESLMGVGTFSADSRGRFPGRYRLAVLENLSRVSSRLRIHGSTG
Mb1630|M.bovis_AF2122/97 LADAELANVLVGVGTFSADSRGQFPGRYRLAVLEKLSRVSSRLRMHGSTG
Rv1604|M.tuberculosis_H37Rv LADAELANVLVGVGTFSADSRGQFPGRYRLAVLEKLSRVSSRLRMHGSTG
MAV_3182|M.avium_104 LTSIDLADALVGAGSFSADARGRFPGRYRMAVLENLSRVSSRLRMHGSTG
Mflv_3608|M.gilvum_PYR-GCK LEPVSLAESIVGTGTFNIDSRGRYPGRYRVKVLENLSRECSRMRMHGATG
Mvan_2809|M.vanbaalenii_PYR-1 LAPTTLAESIVGTGTFNIDSRGRFPGRYRVKVLENLSRECSRMRMHGSTG
MSMEG_3210|M.smegmatis_MC2_155 LGSPTLADSIIGIQTFNIDSRGRFPGRYRVEVLANLSRVCSRVRMHGATG
TH_1158|M.thermoresistible__bu LSDAGLADCVVGTGTFNVDSRGRFPGRYRVALLEALSRRCNRIRMHGATG
MAB_2665c|M.abscessus_ATCC_199 LPRTDLADVAVGLGTFNVDWKGRVPGRYRLTVLEKLSRITSRLRMHGSTG
MLBr_00662|M.leprae_Br4923 LAVSEVAD--LNSASLSFSSLSGWAQRGLRDRFLELTDAVWRVRAYG-DF
* ::: :. ::. . . . * : *: *:* :*
MMAR_2400|M.marinum_M IDLVYVADGILGGAISFGGQVWDHAAGVAQVRAAGGTVTTLTGDPWTPTS
MUL_1577|M.ulcerans_Agy99 IDLVYVADGILGGAISFGGQVWDHAAGVAQVRAAGGTVTTLTGDPWTPTS
Mb1630|M.bovis_AF2122/97 IDLVFVADGILGGAISFGGHVWDHAAGVALVRAAGGVVTDLAGQPWTPAS
Rv1604|M.tuberculosis_H37Rv IDLVFVADGILGGAISFGGHVWDHAAGVALVRAAGGVVTDLAGQPWTPAS
MAV_3182|M.avium_104 LDLAYVADGILGAAVSFGGHVWDHAAGVALVRAAGGVVTDLAGRPWTPAS
Mflv_3608|M.gilvum_PYR-GCK VDFAYVAAGVLGGAISFGHHIWDHAAGVALVRAAGGVVTDLDGEPWTASS
Mvan_2809|M.vanbaalenii_PYR-1 IDLAYVAAGILGGAISFGHHLWDHAAGVALVRAAGGIVTDLAGETWTASS
MSMEG_3210|M.smegmatis_MC2_155 VDLAYVAAGILGGAISFGHHIWDHAAGVALVRAAGGVVTDLTGAPWTVDS
TH_1158|M.thermoresistible__bu VDLAYTAAGILGGAISFGHHIWDHAAGVALIRSAGGVVTDLAGADWTIHS
MAB_2665c|M.abscessus_ATCC_199 IDLAFTAGGILGGAVSFGAHVWDHAAGIALVQAAGGHASDLNGEPWTPES
MLBr_00662|M.leprae_Br4923 LSYCLLAEGAIDVAAEPKVSVWDLAALDIVVREAGGVLTGLDGTPG-PHG
:. * * :. * . :** ** :: *** : * * .
MMAR_2400|M.marinum_M RSVLAAAPGVHEEILEIVRKTGDPEDY--
MUL_1577|M.ulcerans_Agy99 RSVLAAAPGVHEEILEIVRKTGDPEDY--
Mb1630|M.bovis_AF2122/97 RSALAGPLRVHAQILEILGSIGEPEDY--
Rv1604|M.tuberculosis_H37Rv RSALAGPPRVHAQILEILGSIGEPEDY--
MAV_3182|M.avium_104 DSALAAGPGAHAEILDILGNIGRPEDY--
Mflv_3608|M.gilvum_PYR-GCK PSALAAAPGVHEEILQLLRAAGDPKDYL-
Mvan_2809|M.vanbaalenii_PYR-1 SSALAAAPGVHEEILALVRAAGDPKDYL-
MSMEG_3210|M.smegmatis_MC2_155 KSVLAAAPGVHEKMLEIVKSTGKPEDYL-
TH_1158|M.thermoresistible__bu QSVLAGNPGVHAELLEIVQSVGDPEDYRR
MAB_2665c|M.abscessus_ATCC_199 PSLLVAAPGVQEQILEVLAEVGDPEDYR-
MLBr_00662|M.leprae_Br4923 GSAVATNGRLHQEVLTRIGATVK------
* :. : ::* :