For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLVAKTGARLMRSRTLMRAPIWIYRARAGAVFGSRMLMLEHIGRKSGTPRNAVLEVVDHPTPDTYVIASG FGKNSQWFRNIVANPRVRVYVGSHRAAPATARVLEQAEADQALASYRSHHPAAWENLKQVIEETLGSPIT DTDTALPMVELRLD*
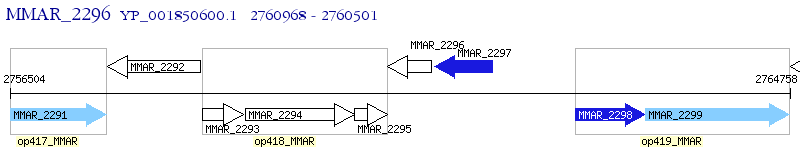
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2296 | - | - | 100% (155) | hypothetical protein MMAR_2296 |
| M. marinum M | MMAR_2750 | - | 3e-05 | 39.51% (81) | hypothetical protein MMAR_2750 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1902c | - | 5e-06 | 40.00% (80) | hypothetical protein Mb1902c |
| M. gilvum PYR-GCK | Mflv_1991 | - | 7e-05 | 32.39% (71) | hypothetical protein Mflv_1991 |
| M. tuberculosis H37Rv | Rv1871c | - | 5e-06 | 40.00% (80) | hypothetical protein Rv1871c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3398 | - | 3e-18 | 38.14% (118) | hypothetical protein MAB_3398 |
| M. avium 104 | MAV_3285 | - | 8e-64 | 73.38% (154) | hypothetical protein MAV_3285 |
| M. smegmatis MC2 155 | MSMEG_1981 | - | 1e-22 | 47.71% (109) | hypothetical protein MSMEG_1981 |
| M. thermoresistible (build 8) | TH_4664 | - | 1e-58 | 70.07% (147) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1499 | - | 2e-79 | 98.61% (144) | hypothetical protein MUL_1499 |
| M. vanbaalenii PYR-1 | Mvan_0980 | - | 3e-45 | 54.90% (153) | hypothetical protein Mvan_0980 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2296|M.marinum_M --------------------------------------------------
MUL_1499|M.ulcerans_Agy99 --------------------------------------------------
MAV_3285|M.avium_104 --------------------------------------------------
TH_4664|M.thermoresistible__bu MTASVVIDPRFHDAVLFELGAVVDAAAGDGAGARVSDSAIILADKLRNAG
Mvan_0980|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3398|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1981|M.smegmatis_MC2_155 --------------------------------------------------
Mb1902c|M.bovis_AF2122/97 --------------------------------------------------
Rv1871c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1991|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2296|M.marinum_M -------------------------------------MSLVAKTGARLMR
MUL_1499|M.ulcerans_Agy99 ------------------------------------------------MR
MAV_3285|M.avium_104 ----------------------MGGVSAAR------ASSLSASIAARLLR
TH_4664|M.thermoresistible__bu IAVAVFAPDGDGXXXAADRHPTKGAIMSSRKYPGKSARNALTDLGARILR
Mvan_0980|M.vanbaalenii_PYR-1 ----------------------------------MAVKSWIANITDRVLH
MAB_3398|M.abscessus_ATCC_1997 -------------------------------------MKKIQKPKPPTGF
MSMEG_1981|M.smegmatis_MC2_155 -------------------------------------MKITKRVHPPSGL
Mb1902c|M.bovis_AF2122/97 -------------------------------------------MNAAMNL
Rv1871c|M.tuberculosis_H37Rv -------------------------------------------MNAAMNL
Mflv_1991|M.gilvum_PYR-GCK ---------------------------------------MPRRVAEFNRR
MMAR_2296|M.marinum_M SRTLMRAPIWIYRARAGAVFGSRMLMLEHIGRKSGTPRNAVLEVVDHP-T
MUL_1499|M.ulcerans_Agy99 SRTLMRAPIWIYRARAGAVFGSRMLMLEHIGRKSGTPRNAVLEVVDHP-A
MAV_3285|M.avium_104 SRQLMRAPIWIYRARGGAVFGSRILLLEHVGRKSGAPRYAALEVVDHP-S
TH_4664|M.thermoresistible__bu SRRLMRAPIWLYRAGAGALLGPRMLMLEHIGRRSGAPRHVVLEVVDHP-E
Mvan_0980|M.vanbaalenii_PYR-1 TRALVRAPIWVYRAGLGFVFGSRLLMLEHIGRTTGARRYVVLEVIDHP-S
MAB_3398|M.abscessus_ATCC_1997 QRLLWRLPITLFRAHLGFLVTKRIMLLTHTGRASGRPRQAALEVVEHG-D
MSMEG_1981|M.smegmatis_MC2_155 SKLLFRAPISMFRANLGWMFGNRLLLLHHIGRVSGKNREVVLEVVEHDPQ
Mb1902c|M.bovis_AF2122/97 KREFVHRVQRFVVNPIGRQLP--MTMLETIGRKTGQPRRTAVGGRVVD--
Rv1871c|M.tuberculosis_H37Rv KREFVHRVQRFVVNPIGRQLP--MTMLETIGRKTGQPRRTAVGGRVVD--
Mflv_1991|M.gilvum_PYR-GCK VTNPATKSLTPWLPNLG--------TLEHVGRKSARRYRTPLLVFETA--
* * ** :. . :
MMAR_2296|M.marinum_M PDTYVIASGFGKNSQWFRNIVANPRVRVYVGSHRAAPATARVLEQAEADQ
MUL_1499|M.ulcerans_Agy99 PDTYVIASGFGKNSQWYRNIVANPRVRVYVGSHRAAPATARVLEQAEADQ
MAV_3285|M.avium_104 PDAYVVASGFGRKAQWFRNIRANPRVRVYAGSHPPRAATARVLDQAEADR
TH_4664|M.thermoresistible__bu PDVYVVASGFGTKAQWFRNITANPQVRVFLGSRAPAQATARVLDRQAADR
Mvan_0980|M.vanbaalenii_PYR-1 PDVYVIASGFGAQAQWFRNVMADARVRISVGGRRSVAATARRLSPGEADL
MAB_3398|M.abscessus_ATCC_1997 DGSIVAASGFGRAADWYKNVRKTPEVTIQVG-GRAHRARAAELSTDEGGD
MSMEG_1981|M.smegmatis_MC2_155 TGAYTVASGWGPKAAWYRNVLAEPRVTIQVG-RRTIAVTALPLDQAEGAD
Mb1902c|M.bovis_AF2122/97 -NQFWMVSEHGEHSDYVYNIKANPAVRVRIG-GRWRSGTAYLLPDDDPRQ
Rv1871c|M.tuberculosis_H37Rv -NQFWMVSEHGEHSDYVYNIKANPAVRVRIG-GRWRSGTAYLLPDDDPRQ
Mflv_1991|M.gilvum_PYR-GCK -DGYAILIGYGPQTDWVKNVLAGGPTVLHKRGRAVVLTDPRVVSKAEAAP
. * : : *: . : . :
MMAR_2296|M.marinum_M ALASYRSHHP----AAWENLKQVIEETLGSPITDTDTALPMVELRLD---
MUL_1499|M.ulcerans_Agy99 ALASYRSHHP----AAWENLKQVIEETLGSPITDTDTALPMVELRLD---
MAV_3285|M.avium_104 ALAAYRTRHP----RAWQRMRPVLEETLGVPITDTGTPLPMVELKLDQRM
TH_4664|M.thermoresistible__bu VLAGYRARHP----RAWSRMKGVIEQTLGEPITDTDTPLPMVELQLRSAA
Mvan_0980|M.vanbaalenii_PYR-1 ALSRYIERHP----RAWESLRTVLEGALQGRVAPPGTEVPMVEIRVR---
MAB_3398|M.abscessus_ATCC_1997 LMAHYAMRRPRSAKQLS-RLMGFEVDGSADDFREVGRAIPFVRFTP-MP-
MSMEG_1981|M.smegmatis_MC2_155 IFARYAVRHRFLARHLLPRLMGFSVNGSLEDFRAAGEQIPFIRFVPRTPG
Mb1902c|M.bovis_AF2122/97 RLRGLPRLNS-------------------AGVRAMGTDLLTIRVDLD---
Rv1871c|M.tuberculosis_H37Rv RLRGLPRLNS-------------------AGVRAMGTDLLTIRVDLD---
Mflv_1991|M.gilvum_PYR-GCK LVVARARPFY-------------------RLFPYDEGALLLTDAGPAR--
. . :
MMAR_2296|M.marinum_M --------
MUL_1499|M.ulcerans_Agy99 --------
MAV_3285|M.avium_104 HTKKPSTP
TH_4664|M.thermoresistible__bu P-------
Mvan_0980|M.vanbaalenii_PYR-1 --------
MAB_3398|M.abscessus_ATCC_1997 --------
MSMEG_1981|M.smegmatis_MC2_155 --------
Mb1902c|M.bovis_AF2122/97 --------
Rv1871c|M.tuberculosis_H37Rv --------
Mflv_1991|M.gilvum_PYR-GCK --------