For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLITGFPAGVLQCNCYVLAERPGTDAIIVDPGQRAMGQLRRILDKNRLTPAAVLITHGHIDHMWSAQKVS DTFGCPTYIHPEDRFMLKDPIYGLGPRVAQLFAGAFFREPKQVVELDRDGDKIDLGNICVNVDHTPGHTR GSVCFRVPQAAKDDRDVVFTGDTLFERSIGRSDLFGGSGRDLLRSIVDKLLVLDDKTVVLPGHGNSTSIG AERRFNPFLEGLST
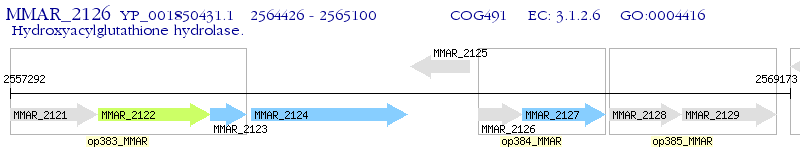
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2126 | - | - | 100% (224) | Zn-dependent glyoxylase |
| M. marinum M | MMAR_3351 | - | 3e-17 | 29.63% (189) | hypothetical protein MMAR_3351 |
| M. marinum M | MMAR_2442 | - | 4e-15 | 27.27% (209) | hypothetical protein MMAR_2442 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2612c | - | 1e-116 | 87.89% (223) | glyoxalase II |
| M. gilvum PYR-GCK | Mflv_3810 | - | 1e-89 | 71.24% (226) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv2581c | - | 1e-116 | 87.89% (223) | glyoxalase II |
| M. leprae Br4923 | MLBr_00493 | - | 1e-111 | 84.68% (222) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_2873c | - | 2e-72 | 57.85% (223) | hypothetical protein MAB_2873c |
| M. avium 104 | MAV_3461 | - | 1e-112 | 85.14% (222) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_2975 | - | 5e-91 | 71.81% (227) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_2332 | - | 1e-94 | 72.07% (222) | POSIBLE GLYOXALASE II (HYDROXYACYLGLUTATHIONE HYDROLASE) (GLX |
| M. ulcerans Agy99 | MUL_1731 | - | 1e-131 | 99.55% (224) | Zn-dependent glyoxylase |
| M. vanbaalenii PYR-1 | Mvan_2589 | - | 1e-89 | 70.93% (227) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2126|M.marinum_M MLITGFPAGVLQCNCYVLAERPGTDAIIVDPGQRAMGQLRRILDKNRLTP
MUL_1731|M.ulcerans_Agy99 MLITGFPAGVLQCNCYALAERPGTDAIIVDPGQRAMGQLRRILDKNRLTP
Mb2612c|M.bovis_AF2122/97 MLITGFPAGLLACNCYVLAERPGTDAVIVDPGQGAMGTLRRILDKNRLTP
Rv2581c|M.tuberculosis_H37Rv MLITGFPAGLLACNCYVLAERPGTDAVIVDPGQGAMGTLRRILDKNRLTP
MLBr_00493|M.leprae_Br4923 MLITGFPAGMLQCNCYVLADRAGTDAVIVDPGQRAMGPLRRILDENRLTP
MAV_3461|M.avium_104 MLITGFPAGMLACNCYVLAQRPGTDAVIVDPGQRAMGPLRRILDRHRLTP
MSMEG_2975|M.smegmatis_MC2_155 MLITGFPAGMLACNCYVLAERPGSDAIVVDPGQRAMGPLREILDEHRLTP
TH_2332|M.thermoresistible__bu VLVTGFPAGMLQCNCYVLAPRRGADAIVIDPGQRAMGPLRRILDENRLTP
Mflv_3810|M.gilvum_PYR-GCK MLITGFPAGLLACNCYVLAPRQGADAIVVDPGQRAMGSLARILDENRLTP
Mvan_2589|M.vanbaalenii_PYR-1 MLITGFPAGMLACNCYVLALRQGADAIVVDPGQRAMGPLRQILDDNRLTP
MAB_2873c|M.abscessus_ATCC_199 MLITGFPAGMFATNCYIVAPHEGGPAVIVDPGQDAIGGVKEILAERDLTP
:*:******:: *** :* : * *:::**** *:* : .** . ***
MMAR_2126|M.marinum_M AAVLITHGHIDHMWSAQKVSDTFGCPTYIHPEDRFMLKDPIYGLGP----
MUL_1731|M.ulcerans_Agy99 AAVLITHGHIDHMWSAQKVSDTFGCPTYIHPEDRFMLKDPIYGLGP----
Mb2612c|M.bovis_AF2122/97 AAVLLTHGHIDHIWSAQKVSDTFGCPTYVHPADRFMLTDPIYGLGP----
Rv2581c|M.tuberculosis_H37Rv AAVLLTHGHIDHIWSAQKVSDTFGCPTYVHPADRFMLTDPIYGLGP----
MLBr_00493|M.leprae_Br4923 SAVLLTHGHIDHMWSAQKVSDTYGCPTYIHPEDRFMLTDPLFGFGP----
MAV_3461|M.avium_104 AAVLLTHGHIDHIWSAQKVSDTYGCPTYIHPEDRFMLKDPIYGLGP----
MSMEG_2975|M.smegmatis_MC2_155 AAVLLTHGHIDHIWSAQKVADTYGCPAYIHPEDRFMLTDPVKAFGRGLIG
TH_2332|M.thermoresistible__bu AAVLLTHGHVDHIWSAQKVADTYGCPAYIHPEDRFMLTDPIKGFGP----
Mflv_3810|M.gilvum_PYR-GCK AAVLLTHGHIDHMWSAQKVADTYGCPTFVHPADRHMLTDPIAGLGQGFLA
Mvan_2589|M.vanbaalenii_PYR-1 AAVLLTHGHIDHMWSAQKVADTYGCPTFIHPADRHMLADPIKGFGRGFLG
MAB_2873c|M.abscessus_ATCC_199 EAVLLTHGHLDHTWTAQPLADEYGIPVYIHPDDRVMLADPLAGIGP----
***:****:** *:** ::* :* *.::** ** ** **: .:*
MMAR_2126|M.marinum_M RVAQLFAGAFFREPKQVVELDRDGDKIDLGNICVNVDHTPGHTRGSVCFR
MUL_1731|M.ulcerans_Agy99 RVAQLFAGAFFREPKQVVELDRDGDKIDLGNICVNVDHTPGHTRGSVCFR
Mb2612c|M.bovis_AF2122/97 RIAQLVAGAFFREPKQVVELDRDGDKIDLGGISVNIDHTPGHTRGSVVFR
Rv2581c|M.tuberculosis_H37Rv RIAQLVAGAFFREPKQVVELDRDGDKIDLGGISVNIDHTPGHTRGSVVFR
MLBr_00493|M.leprae_Br4923 RVAQVVTGAFFREPKQVVELDRDGDKLDLGSVTVNVDHTPGHTRGSVCFW
MAV_3461|M.avium_104 RLAQLAAGAFFREPRQVVELDRDGDKLDLGSVTVNVDHTPGHTRGSMCFR
MSMEG_2975|M.smegmatis_MC2_155 GLGRLAFGALFSEPRQLVELERDGETVGFGGIAVTVDHTPGHTRGSVVFR
TH_2332|M.thermoresistible__bu RIGQLLFGALFAEPRQVIELAVDGEQLNFGELLVTVDHTPGHTRGSVVFR
Mflv_3810|M.gilvum_PYR-GCK GLGRLALSPVFREPRQVLELDRDGDKIELGGITVTVDHTPGHTKGSVVFR
Mvan_2589|M.vanbaalenii_PYR-1 GLARSAVGAMFREPRQVLELDRDGDKIELGGITVAVDHTPGHTQGSVVFR
MAB_2873c|M.abscessus_ATCC_199 GLGQFIQGQVFAEPKELVEIG-DGDKLELAGITLTVDHTPGHTRGSVVFG
:.: . .* **::::*: **: : :. : : :*******:**: *
MMAR_2126|M.marinum_M VPQAAKDD-RDVVFTGDTLFERSIGRSDLFGGSGRDLLRSIVDKLLVLDD
MUL_1731|M.ulcerans_Agy99 VPQAAKDD-RDVVFTGDTLFERSIGRSDLFGGSGRDLLRSIVDKLLVLDD
Mb2612c|M.bovis_AF2122/97 VLQATNND-KDIVFTGDTLFERAIGRTDLAGGSGRDLLRSIVDKLLVLDD
Rv2581c|M.tuberculosis_H37Rv VLQATNND-KDIVFTGDTLFERAIGRTDLAGGSGRDLLRSIVDKLLVLDD
MLBr_00493|M.leprae_Br4923 VAADT-----DVVLTGDTLFERTIGRTDLFGGSGRDLYRSIVEKLLVLDD
MAV_3461|M.avium_104 VSSDK-----QVVFTGDTLFERSVGRTDLFGGSGRDLLTSIVDKLLVLDD
MSMEG_2975|M.smegmatis_MC2_155 VADGD----RDLVFTGDTLFRSSVGRTDLPGGSGRDLLGSIVTKLLVLED
TH_2332|M.thermoresistible__bu VAGDS----TEVAFTGDTLFKQSVGRTDLPGGSGRDLLTSIVNKLMVLDD
Mflv_3810|M.gilvum_PYR-GCK VEQGP----DQVAFTGDTLFRASVGRTDLPGGSGRDLLESIVTKLLVLDD
Mvan_2589|M.vanbaalenii_PYR-1 VEQGT----EPVAFTGDTLFRSSVGRTDLPGGSGRDLMESLVTKLLVLDD
MAB_2873c|M.abscessus_ATCC_199 IEVDSENGPVDVLFSGDTLFQMSIGRTDLPGGNHQQLLDSIAAKLLTRDD
: : ::*****. ::**:** **. ::* *:. **:. :*
MMAR_2126|M.marinum_M KTVVLPGHGNSTSIGAERRFNPFLEGLST
MUL_1731|M.ulcerans_Agy99 KTVVLPGHGNSTSIGAERRFNPFLEGLST
Mb2612c|M.bovis_AF2122/97 STVVLPGHGNSTTIGAERRFNPFLEGLSR
Rv2581c|M.tuberculosis_H37Rv STVVLPGHGNSTTIGAERRFNPFLEGLSR
MLBr_00493|M.leprae_Br4923 KTVVLPGHGNSTTIGAERRFNPFLEGL--
MAV_3461|M.avium_104 DTVVLPGHGNATTIGAERRLNPFLEGL--
MSMEG_2975|M.smegmatis_MC2_155 DAVVLPGHGPKTTIGHERRTNPFLEGLTL
TH_2332|M.thermoresistible__bu DTLVLPGHGATSTIGQERRTNPFLEGLPL
Mflv_3810|M.gilvum_PYR-GCK DTLVLPGHGARSTIGVERHTNPFLEGLKP
Mvan_2589|M.vanbaalenii_PYR-1 DTLVLPGHGERSTIGIERHTNPFLEGLTP
MAB_2873c|M.abscessus_ATCC_199 SSVVLPGHGNATSIGDERRANPFLEGIQ-
.::****** ::** **: ******: