For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPADLHPDLDALAPLLGTWAGQGSGEYPTIEPFEYLEEVVFSHVGKPFLVYAQKTRAVADGAPLHAETGY LRVPKPGQVELVLAHPSGITEIEVGTYSASGGVIEMEMVTTAIGMTPTAKEVTALSRSFRMVGDELSYRL RMGAVGLPLQHHLGARLRRKS
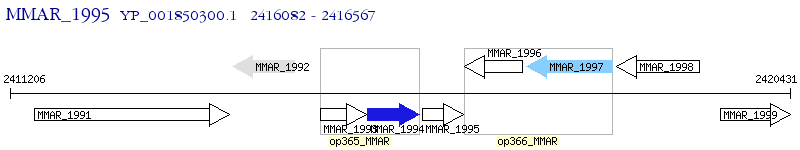
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1995 | - | - | 100% (161) | hypothetical protein MMAR_1995 |
| M. marinum M | MMAR_4871 | - | 2e-10 | 33.33% (171) | hypothetical protein MMAR_4871 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2736c | - | 6e-63 | 72.39% (163) | hypothetical protein Mb2736c |
| M. gilvum PYR-GCK | Mflv_0302 | - | 2e-49 | 54.94% (162) | hypothetical protein Mflv_0302 |
| M. tuberculosis H37Rv | Rv2717c | - | 6e-63 | 72.39% (163) | hypothetical protein Rv2717c |
| M. leprae Br4923 | MLBr_01006 | - | 2e-64 | 70.62% (160) | hypothetical protein MLBr_01006 |
| M. abscessus ATCC 19977 | MAB_0732c | - | 7e-12 | 33.14% (169) | hypothetical protein MAB_0732c |
| M. avium 104 | MAV_3611 | - | 4e-70 | 76.88% (160) | hypothetical protein MAV_3611 |
| M. smegmatis MC2 155 | MSMEG_6574 | - | 1e-58 | 66.67% (156) | hypothetical protein MSMEG_6574 |
| M. thermoresistible (build 8) | TH_2341 | - | 1e-56 | 64.56% (158) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3360 | - | 2e-89 | 98.76% (161) | hypothetical protein MUL_3360 |
| M. vanbaalenii PYR-1 | Mvan_0449 | - | 6e-56 | 64.52% (155) | hypothetical protein Mvan_0449 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0302|M.gilvum_PYR-GCK --------------------------------MVAPVP-LHPDVVALAPL
Mvan_0449|M.vanbaalenii_PYR-1 --------------------------------MADSVPALHPDVAALAPL
TH_2341|M.thermoresistible__bu ------------------------------------VPDLHPDVARLAPL
MSMEG_6574|M.smegmatis_MC2_155 ------------------------------------MIDLHPNIAPLAPL
MMAR_1995|M.marinum_M -----------------------------------MPADLHPDLDALAPL
MUL_3360|M.ulcerans_Agy99 -----------------------------------MPADLHPDLDALAPL
MLBr_01006|M.leprae_Br4923 -----------------------------------MPSDLCPDLQALAPL
MAV_3611|M.avium_104 -----------------------------------MPTDLHPDLAALAPL
Mb2736c|M.bovis_AF2122/97 -----------------------------------MTRDLAPALQALSPL
Rv2717c|M.tuberculosis_H37Rv -----------------------------------MTRDLAPALQALSPL
MAB_0732c|M.abscessus_ATCC_199 MTEPDPAAAEQRPSVGRNLPTFQDLPIPADTANLREGPDLNAAMLALLPL
* . : * **
Mflv_0302|M.gilvum_PYR-GCK LGVWVGEGTGEYPTVPSFAYTEEITFGHVGKPFLSYVQRTRAADDG----
Mvan_0449|M.vanbaalenii_PYR-1 LGTWVGEGSGEYPTIEPFGYTEEITFGHVGKPFLTYAQRTRAADDG----
TH_2341|M.thermoresistible__bu LGTWAGAGTGEYPTIEPFGYYEEVTFDHAGKPFLTYTQRTRAADDG----
MSMEG_6574|M.smegmatis_MC2_155 LGTWAGPGAGEYPTIQSFEYLEEITFGHVGKPFLTYQQRTKARDDG----
MMAR_1995|M.marinum_M LGTWAGQGSGEYPTIEPFEYLEEVVFSHVGKPFLVYAQKTRAVADG----
MUL_3360|M.ulcerans_Agy99 LGTWAGQGAGEYPTIEPFEYLEEVVFSHVGKPFLVYAQKTRAVADG----
MLBr_01006|M.leprae_Br4923 LGSWVGRGMGKYPTIQPFEYLEEVVFSHLDRPFLTYTQKTRAITDG----
MAV_3611|M.avium_104 LGTWTGRGSGKYPTIQPFDYLEEVTFSHVGKPFLAYAQKTRAAADG----
Mb2736c|M.bovis_AF2122/97 LGSWAGRGAGKYPTIRPFEYLEEVVFAHVGKPFLTYTQQTRAVADG----
Rv2717c|M.tuberculosis_H37Rv LGSWAGRGAGKYPTIRPFEYLEEVVFAHVGKPFLTYTQQTRAVADG----
MAB_0732c|M.abscessus_ATCC_199 VGVWRGEGEGRD-TDGDYRFGQQIVVAHDGSDYLTWDARSWRLDADGQFE
:* * * * *. * : : :::.. * . :* : :: .
Mflv_0302|M.gilvum_PYR-GCK RPLHSETGYVRSP-------APHCVEWILAHPTGITEILEGILERIVTGE
Mvan_0449|M.vanbaalenii_PYR-1 RPLHAETGYLRAS-------APDRIEWILAHPTGITEIQEGQLT----AD
TH_2341|M.thermoresistible__bu RPLHTETGYLRVP-------APHRVEWVLAQPTGVTEILEGPLT----VT
MSMEG_6574|M.smegmatis_MC2_155 RPLHAEVGYIRVP-------APDRVEWVLAHPTGITEIQEGTLT----VG
MMAR_1995|M.marinum_M APLHAETGYLRVP-------KPGQVELVLAHPSGITEIEVGTYS----AS
MUL_3360|M.ulcerans_Agy99 TPLHAETGYLRVP-------KPGQVELVLAHPSGITEIEVGTYS----AS
MLBr_01006|M.leprae_Br4923 KPLHAETGYLRVP-------QPGHIELVLAHHSDIAEIEVGTYS----VT
MAV_3611|M.avium_104 KPLHAETGYLRVP-------QPGRLELVLAHPSGITEIEVGSYA----VT
Mb2736c|M.bovis_AF2122/97 KPLHSETGYLRVC-------RPGCVELVLAHPSGITEIEVGTYS----VT
Rv2717c|M.tuberculosis_H37Rv KPLHSETGYLRVC-------RPGCVELVLAHPSGITEIEVGTYS----VT
MAB_0732c|M.abscessus_ATCC_199 QLTLRETGFWRFAPDPNDPDENQAIELVLAHAAGFVELFYGQPLN-----
*.*: * :* :**: :...*: *
Mflv_0302|M.gilvum_PYR-GCK GDTLRIDVES---TAVGRSASAKEVSAVGRTIELA-GDTLTYSVRMAAVG
Mvan_0449|M.vanbaalenii_PYR-1 GDGLRMELVS---SSIGRSGSAKEVTDVGRSIELR-GDTLTYTLRMAAVG
TH_2341|M.thermoresistible__bu DGVIEMELTS---TTVGVTSSAKSVTSVRRSIRVD-GDELTYSLDMAAVG
MSMEG_6574|M.smegmatis_MC2_155 GDRITMDVKA---TTIGLTASAKNVTALARSFRIT-GDELTYTLQMGAVG
MMAR_1995|M.marinum_M GGVIEMEMVT---TAIGMTPTAKEVTALSRSFRMV-GDELSYRLRMGAVG
MUL_3360|M.ulcerans_Agy99 GGVIEMEMVT---TAIGMTPTAKEVTALSRSFRMV-GDELSYRLRMGAVG
MLBr_01006|M.leprae_Br4923 GDLIEVELVT---TTIGLVPTAKQVTALGRFFRID-GDEFAYSVQMGAVG
MAV_3611|M.avium_104 GGLIEMRMST---TSIGLTPTAKEVTALGRWFRID-GDKLSYSVQMGAVG
Mb2736c|M.bovis_AF2122/97 GDVIELELSTRADGSIGLAPTAKEVTALDRSYRID-GDELSYSLQMRAVG
Rv2717c|M.tuberculosis_H37Rv GDVIELELSTRADGSIGLAPTAKEVTALDRSYRID-GDELSYSLQMRAVG
MAB_0732c|M.abscessus_ATCC_199 --ASSWELVT---DALARSKSGALIGGAKRLYGIVDGGDLAYVEERVGAD
: : ::. :. : * : *. ::* ...
Mflv_0302|M.gilvum_PYR-GCK MPLQHHLSAVLHRA--
Mvan_0449|M.vanbaalenii_PYR-1 QPLQHHLSAVLRRVR-
TH_2341|M.thermoresistible__bu HPLQHHLAATLHRKTA
MSMEG_6574|M.smegmatis_MC2_155 QPLQHHLAATLRRTPA
MMAR_1995|M.marinum_M LPLQHHLGARLRRKS-
MUL_3360|M.ulcerans_Agy99 LPLQHHLGARLRRKS-
MLBr_01006|M.leprae_Br4923 QPLQDHLVAVLHRKQ-
MAV_3611|M.avium_104 QPLQDHLAAVLHRQR-
Mb2736c|M.bovis_AF2122/97 QPLQDHLAAVLHRQR-
Rv2717c|M.tuberculosis_H37Rv QPLQDHLAAVLHRQR-
MAB_0732c|M.abscessus_ATCC_199 GGLEPHLSARLSRFAG
*: ** * * *