For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDSSDTTVDGASDGASDGASGADNRAQLVDTALTERQRTILNVIRTSVNDRGYPPSIREIGDAVGLTSTS SVAHQLRTLERKGYLRRDPNRPRAVDVRGADDTVTAAPVTDVAGSDALPEPTFVPVLGRIAAGGPILAEE AVEDVFPLPRELVGQGTLFLLKVVGESMIEAAICDGDWVVVRQQNVADNGDIVAAMIDGEATVKTFKRAG GQIWLMPHNPAFDPIPGNDATVLGKVVTVIRKI*
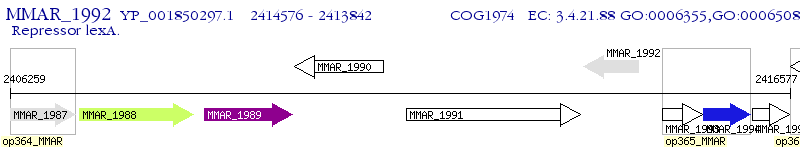
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1992 | lexA | - | 100% (244) | repressor LexA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2739 | lexA | 1e-110 | 91.12% (214) | LexA repressor |
| M. gilvum PYR-GCK | Mflv_3956 | - | 1e-107 | 83.62% (232) | LexA repressor |
| M. tuberculosis H37Rv | Rv2720 | lexA | 1e-111 | 91.59% (214) | LexA repressor |
| M. leprae Br4923 | MLBr_01003 | lexA | 1e-106 | 80.77% (234) | LexA repressor |
| M. abscessus ATCC 19977 | MAB_3038 | - | 1e-105 | 86.79% (212) | LexA repressor |
| M. avium 104 | MAV_3614 | lexA | 1e-114 | 87.88% (231) | LexA repressor |
| M. smegmatis MC2 155 | MSMEG_2740 | lexA | 1e-111 | 83.27% (245) | LexA repressor |
| M. thermoresistible (build 8) | TH_1522 | lexA | 1e-108 | 80.33% (244) | REPRESSOR LEXA |
| M. ulcerans Agy99 | MUL_3363 | lexA | 1e-137 | 99.59% (244) | LexA repressor |
| M. vanbaalenii PYR-1 | Mvan_2441 | - | 1e-109 | 84.91% (232) | LexA repressor |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2739|M.bovis_AF2122/97 MLS--------------------------ADSALTERQRTILDVIRASVT
Rv2720|M.tuberculosis_H37Rv MLS--------------------------ADSALTERQRTILDVIRASVT
MMAR_1992|M.marinum_M MSDSSDTTVDGASDGASDGASGADNRAQLVDTALTERQRTILNVIRTSVN
MUL_3363|M.ulcerans_Agy99 MSDSSDTTVDGASDGASDGASGADNRAQLVDTALTERQRTILNVIRTSVN
MSMEG_2740|M.smegmatis_MC2_155 MSDDTGEFTD--GSTESPADADGAGRRRAVDNGLTERQRTILEVIRASVT
TH_1522|M.thermoresistible__bu MSD----------SHDSPGAADTAGRRRAADSGLTERQRTILEVIRASVT
Mflv_3956|M.gilvum_PYR-GCK MSD------------DSSDSTDAPGTSRSRDSGLTERQRTILDVIRASVT
Mvan_2441|M.vanbaalenii_PYR-1 MSD------------DSSDSTSGAGSGRGRDSGLTERQRTILDVIRASVT
MAB_3038|M.abscessus_ATCC_1997 MSD-------------TPSKGPATGS-------LTERQRTILEVIRASVN
MAV_3614|M.avium_104 MDD-----------SNDSSSAGPDGRLHAVDPSLTERQRTILNVIRASVT
MLBr_01003|M.leprae_Br4923 MSDS-----------TDISGITVDGRLHSMDSGLTERQRTILNVIRASVT
* . *********:***:**.
Mb2739|M.bovis_AF2122/97 SRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVNVRG
Rv2720|M.tuberculosis_H37Rv SRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVNVRG
MMAR_1992|M.marinum_M DRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRG
MUL_3363|M.ulcerans_Agy99 DRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRG
MSMEG_2740|M.smegmatis_MC2_155 SRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRA
TH_1522|M.thermoresistible__bu TRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRG
Mflv_3956|M.gilvum_PYR-GCK TRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDANRPRAVDVRA
Mvan_2441|M.vanbaalenii_PYR-1 SRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRG
MAB_3038|M.abscessus_ATCC_1997 ERGYPPSIREIGDAVGLTSTSSVAHQLRTLEQKGFLRRDPNRPRAVDVRG
MAV_3614|M.avium_104 SRGYPPSIREIGDAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVDVRG
MLBr_01003|M.leprae_Br4923 SRGYPPSIREIADAVGLTSTSSVAHQLRTLERKGYLRRDPNRPRAVNVRG
**********.*******************:**:****.******:**.
Mb2739|M.bovis_AF2122/97 ADD--AALPPVTE--VAGSDALPEPTFAPVLGRIAAGGPILAEEAVEDVF
Rv2720|M.tuberculosis_H37Rv ADD--AALPPVTE--VAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
MMAR_1992|M.marinum_M ADD--TVTAAPVT-DVAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
MUL_3363|M.ulcerans_Agy99 ADD--TVTAAPVT-DVAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
MSMEG_2740|M.smegmatis_MC2_155 ADD--PAAAAVVTTDVAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
TH_1522|M.thermoresistible__bu VDD--PAKPAVTT-DVAGSDTLPEPTFVPVLGRIAAGGPILAEEAVEDVF
Mflv_3956|M.gilvum_PYR-GCK ADD--HPTPIVAT-EVAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
Mvan_2441|M.vanbaalenii_PYR-1 SDD--HAAPIVAT-DVAGSDSLPEPTFVPVLGRIAAGGPILAEEAVEDVF
MAB_3038|M.abscessus_ATCC_1997 IDD--AGTPSATT-DVIGSGDLPEPTFVPVLGRIAAGGPILAEEAVEDVF
MAV_3614|M.avium_104 VDD--DVAAPATE--VAGSDALPEPTFVPVLGRIAAGGPILAEEAVEDVF
MLBr_01003|M.leprae_Br4923 VEETQAAGPAVLT-EVAGSDVLPEPTFVPILGRIAAGSPIFAEGTVEDIF
:: . * **. ******.*:*******.**:** :***:*
Mb2739|M.bovis_AF2122/97 PLPRELVGEGTLFLLKVIGDSMVEAAICDGDWVVVRQQNVADNGDIVAAM
Rv2720|M.tuberculosis_H37Rv PLPRELVGEGTLFLLKVIGDSMVEAAICDGDWVVVRQQNVADNGDIVAAM
MMAR_1992|M.marinum_M PLPRELVGQGTLFLLKVVGESMIEAAICDGDWVVVRQQNVADNGDIVAAM
MUL_3363|M.ulcerans_Agy99 PLPRELVGQGTLFLLKVVGESMVEAAICDGDWVVVRQQNVADNGDIVAAM
MSMEG_2740|M.smegmatis_MC2_155 PLPRELVGEGSLFLLKVVGDSMVDAAICDGDWVVVRQQNVADNGDIVAAM
TH_1522|M.thermoresistible__bu PLPRELVGEGSLFLLKVVGDSMIDAAICDGDWVVVRQQNVAENGDIVAAM
Mflv_3956|M.gilvum_PYR-GCK PLPRELVGEGSLFLLKVVGESMVDAAICDGDWVVVRQQSVADNGDIVAAM
Mvan_2441|M.vanbaalenii_PYR-1 PLPRELVGEGSLFLLKVVGESMIDAAICDGDWVVVRQQSVADNGDIVAAM
MAB_3038|M.abscessus_ATCC_1997 PLPRELVGEGSLFLLKVVGESMVDAAICDGDWVVVRQQNVADNGDIVAAM
MAV_3614|M.avium_104 PLPRELVGDGTLFLLKVVGDSMVEAAICDGDWVVVRQQHVADNGDIVAAM
MLBr_01003|M.leprae_Br4923 PLPRELVGEGTLFLLKVTGDSMVEAAICDGDWVVVRQQKVADNGDIVAAM
********:*:****** *:**::************** **:********
Mb2739|M.bovis_AF2122/97 IDGEATVKTFKRAGGQVWLMPHNPAFDPIPGNDATVLGKVVTVIRKV
Rv2720|M.tuberculosis_H37Rv IDGEATVKTFKRAGGQVWLMPHNPAFDPIPGNDATVLGKVVTVIRKV
MMAR_1992|M.marinum_M IDGEATVKTFKRAGGQIWLMPHNPAFDPIPGNDATVLGKVVTVIRKI
MUL_3363|M.ulcerans_Agy99 IDGEATVKTFKRAGGQIWLMPHNPAFDPIPGNDATVLGKVVTVIRKI
MSMEG_2740|M.smegmatis_MC2_155 IDGEATVKTFKRARGQVWLMPHNPAYDPIPGNEAAVLGKVVTVIRKI
TH_1522|M.thermoresistible__bu IDGEATVKTFKRSRGQVWLMPHNPVYDPIPGNEAAILGKVVTVIRKI
Mflv_3956|M.gilvum_PYR-GCK IDGEATVKTFKRTKGQVWLMPHNPAFDPIPGNDAAILGKVVTVIRKI
Mvan_2441|M.vanbaalenii_PYR-1 IDGEATVKTFKRTKGQVWLMPHNPAFDPIPGNDAAILGKVVTVIRKI
MAB_3038|M.abscessus_ATCC_1997 IDGEATVKTFKRTSGQVWLMPHNPLFEPIPGNDAAILGKVVTVIRKV
MAV_3614|M.avium_104 IDGEATVKTFKRAGGQVWLMPHNPAFDPIPGNDATVLGKVVTVIRKV
MLBr_01003|M.leprae_Br4923 IDGEATVKTFKRAGGQVWLIPHNPAFDPIPGNDATVLGKVVTVIRKI
************: **:**:**** ::*****:*::**********: