For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEALRRVWAKDLDVPTLYELLKLRVEVFVVEQACPYPELDGRDLLAETRHFWLETADGEVICTLRLMEE HAGGEKVFRLGRLCTKRQARGHGHSTRLLRAALAEVGDYPCRIDAQVYLAEMYARHGFVRDGDDFIDDGI PHLPMLRPAHGPAAQS
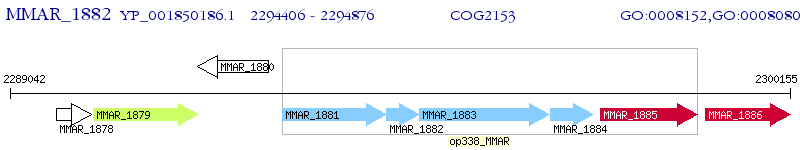
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1882 | elaA | - | 100% (156) | acyltransferase ElaA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2876c | - | 3e-76 | 86.75% (151) | hypothetical protein Mb2876c |
| M. gilvum PYR-GCK | Mflv_4051 | - | 1e-66 | 77.03% (148) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv2851c | - | 4e-76 | 86.75% (151) | hypothetical protein Rv2851c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3158c | - | 7e-38 | 51.82% (137) | hypothetical protein MAB_3158c |
| M. avium 104 | MAV_3707 | - | 2e-77 | 86.27% (153) | acetyltransferase, gnat family protein |
| M. smegmatis MC2 155 | MSMEG_2614 | - | 3e-69 | 79.73% (148) | ElaA protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2110 | elaA | 2e-77 | 99.26% (136) | acyltransferase ElaA |
| M. vanbaalenii PYR-1 | Mvan_2301 | - | 2e-67 | 78.38% (148) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4051|M.gilvum_PYR-GCK MTVALRRSWAKDLSAATLYELLKLRVEVFVVEQATPYPELDGRDLLAETR
Mvan_2301|M.vanbaalenii_PYR-1 MTVALRRSWAKDLDAATLYELLKLRVEVFVVEQATPYPELDGRDLLAETR
MSMEG_2614|M.smegmatis_MC2_155 MTVSLRRSWAKDLDAPTLYELLKLRVEVFVVEQATPYPELDGRDLLAETR
MMAR_1882|M.marinum_M MSEALRRVWAKDLDVPTLYELLKLRVEVFVVEQACPYPELDGRDLLAETR
MUL_2110|M.ulcerans_Agy99 --------------------MLKLRVEVFVVEQACRYPELDGRDLLAETR
Mb2876c|M.bovis_AF2122/97 MTEALRRVWAKDLDARALYELLKLRVEVFVVEQACPYPELDGRDLLAETR
Rv2851c|M.tuberculosis_H37Rv MTEALRRVWAKDLDARALYELLKLRVEVFVVEQACPYPELDGRDLLAETR
MAV_3707|M.avium_104 MTEALRRTWAKDLDAQTLYELLKLRVEVFVVEQAIPYPELDGRDLLAETR
MAB_3158c|M.abscessus_ATCC_199 --MSIGWSWSADLDTTTLYGLLRLRAEAFIVGQNCAYQDLDGRDLDPGTR
:*:**.*.*:* * * :****** . **
Mflv_4051|M.gilvum_PYR-GCK HFWLENPEGEVISTLRLMEEHPAGQKAFRIGRVCTKRTARGHGHTARLLT
Mvan_2301|M.vanbaalenii_PYR-1 HFWLENPEGEVISTLRLMEEHPAGQKAFRIGRVCTKRGDRGNGHTARLLQ
MSMEG_2614|M.smegmatis_MC2_155 HFWLEGPDGEVISTLRLMEEHPGGEKVFRIGRVCTKRSARGQGHVARLMQ
MMAR_1882|M.marinum_M HFWLETADGEVICTLRLMEEHAGGEKVFRLGRLCTKRQARGHGHSTRLLR
MUL_2110|M.ulcerans_Agy99 HFWLETADGEVICTLRLMEEHAGGEKVFRLGRLCTKRQARGHGHSTRLLR
Mb2876c|M.bovis_AF2122/97 HFWLETPDGEVTCTLRLMEEHAGGEKVFRIGRLCTKRDARGQGHSNRLLC
Rv2851c|M.tuberculosis_H37Rv HFWLETPDGEVTCTLRLMEEHAGGEKVFRIGRLCTKRDARGQGHSNRLLC
MAV_3707|M.avium_104 HFWLETSDGEVICTLRLMEEHAGGEKAFRIGRLCTKRSARGQGHTTRLLR
MAB_3158c|M.abscessus_ATCC_199 HFWIKDQDNAVVSGLRLLVDPAGDG--FWIGRVCTAEAERGRGHAARLVR
***:: :. * . ***: : ... * :**:** . **.** **:
Mflv_4051|M.gilvum_PYR-GCK AALAEVGDFPCHINAQTYLVQMYGRHGFVRDGAEFLDDGIPHVPMVKP--
Mvan_2301|M.vanbaalenii_PYR-1 AALAEVGDYPCHINAQTYLEEMYRRHGFVRDGDEFLDDGIPHVPMVKP--
MSMEG_2614|M.smegmatis_MC2_155 AALAEVGDYPCHINAQTYLEDMYGKHGFVRDGEEFLDDGIPHIPMVRPGT
MMAR_1882|M.marinum_M AALAEVGDYPCRIDAQVYLAEMYARHGFVRDGDDFIDDGIPHLPMLRPAH
MUL_2110|M.ulcerans_Agy99 AALAEVGDYPCRIDAQVYLAEMYARHGFVRDGDDFIDDGIPHLPMLRPAH
Mb2876c|M.bovis_AF2122/97 AALAEVGDYPCRIDAQAYLTAMYAQHGFVRDGDEFLDDGIPHVPMLRPGS
Rv2851c|M.tuberculosis_H37Rv AALAEVGDYPCRIDAQAYLTAMYAQHGFVRDGDEFLDDGIPHVPMLRPGS
MAV_3707|M.avium_104 AALAEVGDYPCRINAQTYLADMYAQHGFVRDGDDFLDDGVPHVPMLRPGS
MAB_3158c|M.abscessus_ATCC_199 AVLAEIGGAGCRLNAQAHLKDMYGKLGFVVSGDEFLEDEIPHVPMTWVAG
*.***:*. *:::**.:* ** : *** .* :*::* :**:**
Mflv_4051|M.gilvum_PYR-GCK -------
Mvan_2301|M.vanbaalenii_PYR-1 -------
MSMEG_2614|M.smegmatis_MC2_155 RAEAEQP
MMAR_1882|M.marinum_M GPAAQS-
MUL_2110|M.ulcerans_Agy99 GPAAQS-
Mb2876c|M.bovis_AF2122/97 GQVERP-
Rv2851c|M.tuberculosis_H37Rv GQVERP-
MAV_3707|M.avium_104 GLAEKP-
MAB_3158c|M.abscessus_ATCC_199 NG-----