For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTAIDRGLSSELAADLAAAAFTLAKRFAAGATMWSIAPSWEPHALHIAVEFVHPVIMGKRALPAVALTG PDLVDLVRVSVRPGDIVVAVAGADQPDVRSVMRRSPAWGATTIWIGSGARPEAALADHLLWLDDPDPRVP ATGGFVLFYHLLWELTHVCFEHSGLLKPECTEEVCVTCSDEGRLGEVVSAPADGLAAVRTARGIEDVITT LIDPVAAGDLVLVHAGTAISRLGDDGDDS*
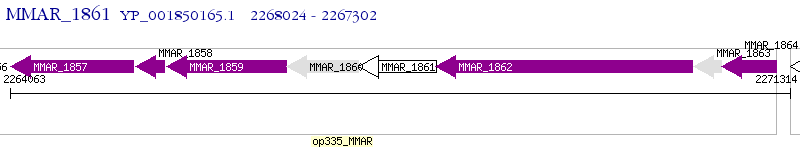
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1861 | - | - | 100% (240) | hypothetical protein MMAR_1861 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2707 | - | 1e-73 | 59.40% (234) | hypothetical protein MSMEG_2707 |
| M. thermoresistible (build 8) | TH_0223 | - | 3e-98 | 69.11% (246) | hypothetical protein MSMEG_2707 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1861|M.marinum_M MTTTAIDRGLSSELAADLAAAAFTLAKRFAAGATMWSIAPSWEPHALHIA
TH_0223|M.thermoresistible__bu VITTAVDRGLDADLAADLAAAALTLARRFSAEATMWCLAPGWEPHAQHVA
MSMEG_2707|M.smegmatis_MC2_155 MTTIPAPTFVGTDLSADLATAALDVARKFHDGATLWVIAPQWEPHAHHVA
: * . :.::*:****:**: :*::* **:* :** ***** *:*
MMAR_1861|M.marinum_M VEFVHPVIMGKRALPAVALTGPDLVDLVRVSVRPGDIVVAVAGADQPDVR
TH_0223|M.thermoresistible__bu VEFVHPVIVGKRALPAVAVTGADPVGALRMSVRPGDIVLAVCGADHPDVR
MSMEG_2707|M.smegmatis_MC2_155 VEFVHPVIMGKRALPSVALVESDPVAQARVASQPGDLLLAVAAADDAAVI
********:******:**:. .* * *:: :***:::**..**.. *
MMAR_1861|M.marinum_M SVMRRSPAWGATTIWIGSGARPEAALADHLLWLDDPDPRVPATGGFVLFY
TH_0223|M.thermoresistible__bu SAMLRAPAWGATTVWIGHGARPPAGAADHVLWLDDPDPRIPATGGFVLLY
MSMEG_2707|M.smegmatis_MC2_155 DAMRRADAWGVTTLWIGSGPRPPAGAAHHILWVDSDDPMVPATGRFVLMY
..* *: ***.**:*** *.** *. *.*:**:*. ** :**** ***:*
MMAR_1861|M.marinum_M HLLWELTHVCFEHSGLLK-PECTEEVCVTCSDEGRLGEVVSAPAD-----
TH_0223|M.thermoresistible__bu HLLWELTHVCFEHPGLLREPECTDEVCITCSDDGCLAEVIDSPGSPGCPG
MSMEG_2707|M.smegmatis_MC2_155 HLLWELTHVCFEHPGLLTTPADDTHTCVTCSDEGRLAEVILPPAGAG---
*************.*** * ..*:****:* *.**: .*..
MMAR_1861|M.marinum_M --GLAAVRTARGIEDVITTLIDPVAAGDLVLVHAGTAISRLGDDGDDS--
TH_0223|M.thermoresistible__bu GVGGVVARTAEGTVTVDTSLVGPVDAGDLLLVHAGLAISKVTDDTRDAVT
MSMEG_2707|M.smegmatis_MC2_155 --REALVRTARGEEWVDTAMLGAVDVDDLLLVHGGVAIAAVTAAEDEVTP
. .***.* * *:::..* ..**:***.* **: : :
MMAR_1861|M.marinum_M -------
TH_0223|M.thermoresistible__bu GDGEVGR
MSMEG_2707|M.smegmatis_MC2_155 -------