For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIDEALFDAEEKMEKAVSVAREDMATIRTGRANPGMFSRIVIDYYGTHTPITQLASINVPEARMVVIKPY EANQLHAIETAIRNSDLGVNPTNDGTIIRVAVPQLTEERRRDLVKQAKHKGEEARVSVRNIRRKAMEELH RIRKDGEAGEDEVARAEKDLDKSTHQYVAQIDELVKHKEGELLEV
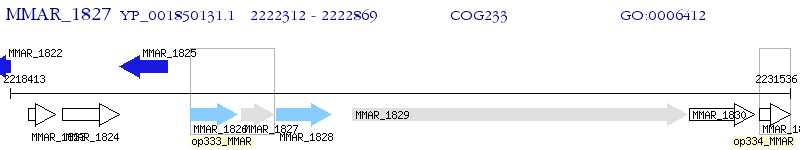
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1827 | frr | - | 100% (185) | ribosome recycling factor Frr |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2906c | frr | 2e-93 | 90.81% (185) | ribosome recycling factor |
| M. gilvum PYR-GCK | Mflv_4123 | frr | 3e-87 | 84.32% (185) | ribosome recycling factor |
| M. tuberculosis H37Rv | Rv2882c | frr | 2e-93 | 90.81% (185) | ribosome recycling factor |
| M. leprae Br4923 | MLBr_01590 | frr | 4e-92 | 89.73% (185) | ribosome recycling factor |
| M. abscessus ATCC 19977 | MAB_3187c | frr | 6e-85 | 82.70% (185) | ribosome recycling factor |
| M. avium 104 | MAV_3732 | frr | 4e-92 | 89.73% (185) | ribosome recycling factor |
| M. smegmatis MC2 155 | MSMEG_2541 | frr | 4e-86 | 84.86% (185) | ribosome recycling factor |
| M. thermoresistible (build 8) | TH_0272 | frr | 5e-85 | 82.70% (185) | RIBOSOME RECYCLING FACTOR FRR (RIBOSOME RELEASING FACTOR) |
| M. ulcerans Agy99 | MUL_2075 | frr | 1e-101 | 99.46% (185) | ribosome recycling factor |
| M. vanbaalenii PYR-1 | Mvan_2221 | frr | 5e-85 | 83.24% (185) | ribosome recycling factor |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1827|M.marinum_M MIDEALFDAEEKMEKAVSVAREDMATIRTGRANPGMFSRIVIDYYGTHTP
MUL_2075|M.ulcerans_Agy99 MIDEALFDAEEKMEKAVSVAREDMATIRTGRANPGMFSRIVIDYYGTHTP
Mb2906c|M.bovis_AF2122/97 MIDEALFDAEEKMEKAVAVARDDLSTIRTGRANPGMFSRITIDYYGAATP
Rv2882c|M.tuberculosis_H37Rv MIDEALFDAEEKMEKAVAVARDDLSTIRTGRANPGMFSRITIDYYGAATP
MAV_3732|M.avium_104 MIDEALFDAEEKMEKAVAVARDDLSTIRTGRANPGMFSRIVIDYYGAATP
MLBr_01590|M.leprae_Br4923 MIDEALFDAEEKMEKAVSVAREDLSMIRTGRANPGMFSRLVIDYYGSATP
Mflv_4123|M.gilvum_PYR-GCK MIDETLFDAEEKMEKAVSVARDDLASIRTGRANPGMFNRIHIDYYGASTP
Mvan_2221|M.vanbaalenii_PYR-1 MIDETLFDAEEKMEKAVAVARDDLASIRTGRANPGMFNRIHVDYYGAVTP
MSMEG_2541|M.smegmatis_MC2_155 MIDETLFDAEEKMEKAVSVARDELGSIRTGRANPGMFNRINIDYYGSMTP
TH_0272|M.thermoresistible__bu VIEETLFDAEEKMEKAVDVARDDLASIRTGRANPGMFSRVLIDYYGTPTP
MAB_3187c|M.abscessus_ATCC_199 MIDEVLLDAEEKMEKAVTVARDDFATIRTGRANPGMFQRVVIDYYGTPTP
:*:*.*:********** ***:::. ***********.*: :****: **
MMAR_1827|M.marinum_M ITQLASINVPEARMVVIKPYEANQLHAIETAIRNSDLGVNPTNDGTIIRV
MUL_2075|M.ulcerans_Agy99 ITQLASINVPEARMVVIKPYEANQLHAIETAIRNSDLGVNPTNDGTIIRV
Mb2906c|M.bovis_AF2122/97 ITQLASINVPEARLVVIKPYEANQLRAIETAIRNSDLGVNPTNDGALIRV
Rv2882c|M.tuberculosis_H37Rv ITQLASINVPEARLVVIKPYEANQLRAIETAIRNSDLGVNPTNDGALIRV
MAV_3732|M.avium_104 ITQLASINVPEARLVVIKPYEASQVGAIETAIRNSDLGVNPTNDGTLIRV
MLBr_01590|M.leprae_Br4923 ITQLASINVPEARLVVIKPYDAIQLHAIETAIRNSDLGVNPSNDGTLIRV
Mflv_4123|M.gilvum_PYR-GCK ITQLSSINVPEARMVVIKPYEANQLRPIEDAIRNSDLGVNPTNDGNIIRV
Mvan_2221|M.vanbaalenii_PYR-1 ITQLSSINVPEARMVVIKPYEASQLRPIEDAIRNSDLGVNPTNDGNVIRV
MSMEG_2541|M.smegmatis_MC2_155 ITQLASINVPEARLVVIKPYEASQLRAIEDAIRNSDLGVNPSNDGNIIRV
TH_0272|M.thermoresistible__bu ITQLSSINVPEARLVVIKPYEANQLRNIEEAIRNSDLGLNPTNDGNVIRV
MAB_3187c|M.abscessus_ATCC_199 ITQVAGINVPEARMVVIKPYESNQLKAIEDAIRNSDLGLNPSNDGSIIRV
***::.*******:******:: *: ** ********:**:*** :***
MMAR_1827|M.marinum_M AVPQLTEERRRDLVKQAKHKGEEARVSVRNIRRKAMEELHRIRKDGEAGE
MUL_2075|M.ulcerans_Agy99 AVPQLTEERRRDLVKQAKHKGEEARVSVRNIRRKAMEELHRIRKDGEAGE
Mb2906c|M.bovis_AF2122/97 AVPQLTEERRRELVKQAKHKGEEAKVSVRNIRRKAMEELHRIRKEGEAGE
Rv2882c|M.tuberculosis_H37Rv AVPQLTEERRRELVKQAKHKGEEAKVSVRNIRRKAMEELHRIRKEGEAGE
MAV_3732|M.avium_104 AVPQLTEERRRELVKQAKSKGEDAKVSVRNIRRKAMEELHRIRKDGEAGE
MLBr_01590|M.leprae_Br4923 AVPQLTEERRRELVKQAKCKGEDAKVSVRNIRRKVMEELHRIRKDGEAGE
Mflv_4123|M.gilvum_PYR-GCK SIPQLTEERRRDLVKQAKSKGEDAKVSIRNIRRKAMEELARIKKDGDAGE
Mvan_2221|M.vanbaalenii_PYR-1 SIPQLTEERRRDLVKQAKGKGEDAKVSVRNIRRKAMEELTRIKKDGDAGE
MSMEG_2541|M.smegmatis_MC2_155 AIPQLTEERRRELVKQAKSKGEDAKVSVRNIRRKAMEELSRIKKDGEAGE
TH_0272|M.thermoresistible__bu AIPPLTEERRRELVKQAKAKGEEAKVSIRAVRRKAMEELHRIKKDGEAGE
MAB_3187c|M.abscessus_ATCC_199 AIPQLTEERRRELVKQAKGKGEDAKVTLRNIRRKANDELNRIKKDGEAGE
::* *******:****** ***:*:*::* :***. :** **:*:*:***
MMAR_1827|M.marinum_M DEVARAEKDLDKSTHQYVAQIDELVKHKEGELLEV
MUL_2075|M.ulcerans_Agy99 DEVARAEKDLDKSTHQYVAQIDELVKHKEGDLLEV
Mb2906c|M.bovis_AF2122/97 DEVGRAEKDLDKTTHQYVTQIDELVKHKEGELLEV
Rv2882c|M.tuberculosis_H37Rv DEVGRAEKDLDKTTHQYVTQIDELVKHKEGELLEV
MAV_3732|M.avium_104 DEVGRAEKDLDKTTHQYITQIDELVKHKEGELLEV
MLBr_01590|M.leprae_Br4923 DEVSRAEKDLDKTTHQYVIQIDELVKHKEGELLEV
Mflv_4123|M.gilvum_PYR-GCK DDVTRAEKDLDKSTHQYTSQVDDLVKHKEGELLEV
Mvan_2221|M.vanbaalenii_PYR-1 DDVARAEKDLDKTTQQYTHQIDELVKHKEGELLEV
MSMEG_2541|M.smegmatis_MC2_155 DEVSRAEKDLDKSTHTYTAQIDELVKHKESELLEV
TH_0272|M.thermoresistible__bu DEVARAEKDLDKTTAQYTGQIDELVKTKEGELLEV
MAB_3187c|M.abscessus_ATCC_199 DEVGRAEKDLDKTTAQYVSQIDELVKHKEGELLEV
*:* ********:* * *:*:*** **.:****