For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTMWGAPLHRRWHGSRLRDPRQAKFLTLASLKWVLANRAYTPWYLVRYWRLLKFKLANPHIITRGMVFL GKGVEIHATPELAQLEIGRWVHIGDKNTIRAHEGSLRFGDKVVLGRDNVINCYLDIELGDSALMADWCYV CDFDHRMDDINVPIKDQGIIKSPVRIGPDTWVGVKVTVLRGTSVGRGCVLGAHAVVRGDIPDYSIAVGSP AKVVKNRQLSWETSAAQRAELAAALADIERKKAAH
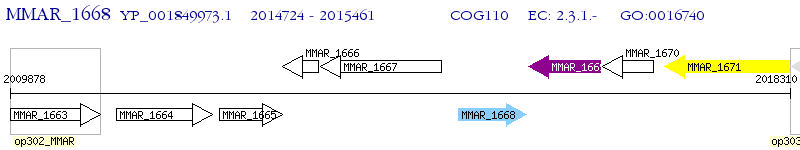
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1668 | - | - | 100% (245) | transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3060c | - | 1e-137 | 93.44% (244) | transferase |
| M. gilvum PYR-GCK | Mflv_4264 | - | 1e-130 | 88.93% (244) | hypothetical protein Mflv_4264 |
| M. tuberculosis H37Rv | Rv3034c | - | 1e-137 | 93.44% (244) | transferase |
| M. leprae Br4923 | MLBr_01719 | - | 1e-138 | 94.29% (245) | hypothetical protein MLBr_01719 |
| M. abscessus ATCC 19977 | MAB_3378c | - | 1e-122 | 81.15% (244) | hypothetical protein MAB_3378c |
| M. avium 104 | MAV_3898 | - | 1e-138 | 95.08% (244) | hexapeptide transferase family protein |
| M. smegmatis MC2 155 | MSMEG_2335 | - | 1e-129 | 87.65% (243) | hexapeptide transferase family protein |
| M. thermoresistible (build 8) | TH_2190 | - | 1e-130 | 88.52% (244) | POSSIBLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_1904 | - | 1e-145 | 100.00% (245) | transferase |
| M. vanbaalenii PYR-1 | Mvan_2095 | - | 1e-130 | 88.89% (243) | hexapaptide repeat-containing transferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4264|M.gilvum_PYR-GCK --------------------------MHGRGPPARGPGEPADPGGVGDGR
Mvan_2095|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2335|M.smegmatis_MC2_155 --------------------------------------------------
TH_2190|M.thermoresistible__bu --------------------------------------------------
MMAR_1668|M.marinum_M --------------------------------------------------
MUL_1904|M.ulcerans_Agy99 --------------------------------------------------
Mb3060c|M.bovis_AF2122/97 MNVLSLGSSSGVVWGRVPITAPAGAATGVTSRADAHSQMRRYAQTGPTAK
Rv3034c|M.tuberculosis_H37Rv MNVLSLGSSSGVVWGRVPITAPAGAATGVTSRADAHSQMRRYAQTGPTAK
MAV_3898|M.avium_104 --------------------------------------------------
MLBr_01719|M.leprae_Br4923 --------------------------------------------------
MAB_3378c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4264|M.gilvum_PYR-GCK VGFSAMTTMWGAPLHKRWRGSRLRDPRQARFLTLASLKWVLKNRAFTPWY
Mvan_2095|M.vanbaalenii_PYR-1 -----MTTMWGAPLHKRWRGSRLKDPRQADFLTLASLKWVIANRAYTPWY
MSMEG_2335|M.smegmatis_MC2_155 -----MTTMWGAPIHKRWRGSRLRDPRQADFLTMASLKWVIANRAFTPWY
TH_2190|M.thermoresistible__bu -----MTTMWGAPLHVRWRGSRLRDPRQARFLTWASLKWVIANRAYTPWY
MMAR_1668|M.marinum_M -----MTTMWGAPLHRRWHGSRLRDPRQAKFLTLASLKWVLANRAYTPWY
MUL_1904|M.ulcerans_Agy99 -----MTTMWGAPLHRRWHGSRLRDPRQAKFLTLASLKWVLANRAYTPWY
Mb3060c|M.bovis_AF2122/97 LSSAPMTTMWGAPLHRRWRGSRLRDPRQAKFLTLASLKWVLANRAYTPWY
Rv3034c|M.tuberculosis_H37Rv LSSAPMTTMWGAPLHRRWRGSRLRDPRQAKFLTLASLKWVLANRAYTPWY
MAV_3898|M.avium_104 -----MTTMWGAPLHRRWRGSNLRDPRQAKFLTLASLKWVLRNRAYTPWY
MLBr_01719|M.leprae_Br4923 -----MTTMWGAPLHRRWCGSRLRDPLQAKFLTLASLKWVLANRAYTPWY
MAB_3378c|M.abscessus_ATCC_199 -----MTTMWGAPLHKRWRGQRLRDPLQAKFLTRDSLRWVIANKAYTPWY
********:* ** *..*:** ** *** **:**: *:*:****
Mflv_4264|M.gilvum_PYR-GCK LVRYFRLLKFKLANPHIITRGMVFLGKDVEIQATPELSQMEIGRWVHIGD
Mvan_2095|M.vanbaalenii_PYR-1 LVRYWRLLKFKLANPHIITRGMVFLGKDVEIQATPELSQMEIGRWVHIGD
MSMEG_2335|M.smegmatis_MC2_155 LVRYWRLLKFKLANPHIITRGMVFLGKDVEIQCTPELAQMEIGRWVHIGD
TH_2190|M.thermoresistible__bu LVRYWRLLKFKLANPHIITRGMVFLGKDVEIHATPELAQLEIGRWVHIGD
MMAR_1668|M.marinum_M LVRYWRLLKFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
MUL_1904|M.ulcerans_Agy99 LVRYWRLLKFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
Mb3060c|M.bovis_AF2122/97 LVRYWRLLRFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
Rv3034c|M.tuberculosis_H37Rv LVRYWRLLRFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
MAV_3898|M.avium_104 LVRYWRLLKFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
MLBr_01719|M.leprae_Br4923 LVRYWRLLKFKLANPHIITRGMVFLGKGVEIHATPELAQLEIGRWVHIGD
MAB_3378c|M.abscessus_ATCC_199 LVRYWRLLKFKLANPHIVTRGMLFLGKNVEIHCTPELARMEIGRWVHIGD
****:***:********:****:****.***:.****:::**********
Mflv_4264|M.gilvum_PYR-GCK KNTIRCHEGSLRIGDKVVLGRDNVINTYLDIELGDSVLMADWCYICDFDH
Mvan_2095|M.vanbaalenii_PYR-1 KNTIRCHEGSLRIGDKVVLGRDNVINTYLDIELGDSVLMADWCYVCDFDH
MSMEG_2335|M.smegmatis_MC2_155 KNTIRCHEGSLRIGDKVVLGRDNVINTYLDIELGDSVLMADWCYVCDFDH
TH_2190|M.thermoresistible__bu KNNIRAHEGSLRIGDKVVLGRDNVINAYLDIELGDSVLMADWCYIVDFDH
MMAR_1668|M.marinum_M KNTIRAHEGSLRFGDKVVLGRDNVINCYLDIELGDSALMADWCYVCDFDH
MUL_1904|M.ulcerans_Agy99 KNTIRAHEGSLRFGDKVVLGRDNVINCYLDIELGDSALMADWCYVCDFDH
Mb3060c|M.bovis_AF2122/97 KNTIRAHEGSLRFGDKVVLGRDNVINTYLDIEIGDSVLMADWCYICDFDH
Rv3034c|M.tuberculosis_H37Rv KNTIRAHEGSLRFGDKVVLGRDNVINTYLDIEIGDSVLMADWCYICDFDH
MAV_3898|M.avium_104 KNTIRAHEGSLRFGDKVVLGRDNVINAYLDIELGDSVLMADWCYVCDFDH
MLBr_01719|M.leprae_Br4923 KNTIRAHEGSLRFGDKVVLGRDNVINCYLDIELGSSALMADWCYVCDFDH
MAB_3378c|M.abscessus_ATCC_199 GNSLRAHEGSLRLGDKVVFGKDNVVNAYIDIEFGDSALMADWCYVCDFDH
*.:*.******:*****:*:***:* *:***:*.*.*******: ****
Mflv_4264|M.gilvum_PYR-GCK RMDSIELPIKDQGIVKGPVRIGPDTWVGVKVSVLRNTSIGRGCVLGSHAV
Mvan_2095|M.vanbaalenii_PYR-1 KMESIELPIKDQGIVKSPVRIGPDTWVAAKVTVLRGTSIGRGCVLGAHAV
MSMEG_2335|M.smegmatis_MC2_155 KMDDINVPIKDQGIIKGPVRIGPDTWIAAKVTILRNTLIGRGCVLGAHAV
TH_2190|M.thermoresistible__bu KMDSIEMPIKDQGIVKSPVRIGPDTWVGAKVSVLRGTSIGRGCVLGSHAV
MMAR_1668|M.marinum_M RMDDINVPIKDQGIIKSPVRIGPDTWVGVKVTVLRGTSVGRGCVLGAHAV
MUL_1904|M.ulcerans_Agy99 RMDDINVPIKDQGIIKSPVRIGPDTWVGVKVTVLRGTSVGRGCVLGAHAV
Mb3060c|M.bovis_AF2122/97 RMDDITLPIKDQGIIKSPVRIGPDTWIGVKVSVLRGTTIGRGCVLGSHAV
Rv3034c|M.tuberculosis_H37Rv RMDDITLPIKDQGIIKSPVRIGPDTWIGVKVSVLRGTTIGRGCVLGSHAV
MAV_3898|M.avium_104 RMDDINVPIKDQGIVKSPVRIGPDTWVGVKVTVLRGTSIGRGCVLGSHAV
MLBr_01719|M.leprae_Br4923 RMDDINIPIKDQGIVKSPVRIGPDTWMAAKVTVLRGTSVGRGCVLGAHAV
MAB_3378c|M.abscessus_ATCC_199 RMEDITYPIKDQGIVKSPVRIGPDTWIAAKVTILRDTIVGRGCVLGAHAV
:*:.* *******:*.*********:..**::**.* :*******:***
Mflv_4264|M.gilvum_PYR-GCK VRGDIPDYSIAVGAPAKVVKNRKLSWETSAAERAELAAALADIERKKAAR
Mvan_2095|M.vanbaalenii_PYR-1 VKGEIPDYSIAVGAPAKVVKNRKLSWETSAAERAELAAALADIERKKAVR
MSMEG_2335|M.smegmatis_MC2_155 VKGDIPDHSIAVGAPAKVVKNRKLAWETSAAERAELAAALADIERKKAVS
TH_2190|M.thermoresistible__bu VRGEIPDYSIAVGAPAKVIKNRRLAWEASAAERAELAAALADIERKKAAR
MMAR_1668|M.marinum_M VRGDIPDYSIAVGSPAKVVKNRQLSWETSAAQRAELAAALADIERKKAAH
MUL_1904|M.ulcerans_Agy99 VRGDIPDYSIAVGSPAKVVKNRQLSWETSAAQRAELAAALADIERKKAAH
Mb3060c|M.bovis_AF2122/97 VRGAIPDYSIAVGAPAKVVKNRQLSWEASAAQRAELAAALADIERKKAAR
Rv3034c|M.tuberculosis_H37Rv VRGAIPDYSIAVGAPAKVVKNRQLSWEASAAQRAELAAALADIERKKAAR
MAV_3898|M.avium_104 VRGVIPDYSIAVGAPAKVVKNRQLSWESSAAQRAELAAALADIERKKASR
MLBr_01719|M.leprae_Br4923 VRGVIPDYSVAVGAPAKVVNNRKLSWEQSAAQRAELAAALADIERKKAAH
MAB_3378c|M.abscessus_ATCC_199 VKGVIPDYSIAVGAPAKVVKNRKVAWDASGAQRAQLAEALADIERKKAAA
*:* ***:*:***:****::**:::*: *.*:**:** **********
Mflv_4264|M.gilvum_PYR-GCK ----
Mvan_2095|M.vanbaalenii_PYR-1 ----
MSMEG_2335|M.smegmatis_MC2_155 NK--
TH_2190|M.thermoresistible__bu ----
MMAR_1668|M.marinum_M ----
MUL_1904|M.ulcerans_Agy99 ----
Mb3060c|M.bovis_AF2122/97 ----
Rv3034c|M.tuberculosis_H37Rv ----
MAV_3898|M.avium_104 ----
MLBr_01719|M.leprae_Br4923 ----
MAB_3378c|M.abscessus_ATCC_199 NSPE