For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDLNYAGVLDLAPGGLVLTAVLIVWRRDLRSIVWLLAAQGLALAAIPMILGVHSHDWELVGIGLALFAL RAVVLPWLLARALGAEKHEQREATPLVNTTASLLIAAVLTVVALAVTRPIVDLDSSAAVSAVPGGLAVVL IALFVMVTRRHAISQAAGFLMLDNGIAATAFLLTAGVPLIVEMGASLDVLFALIVLGVLTGRLRRTFGGT DLDRLQELRD
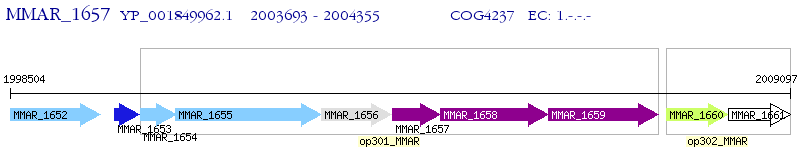
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1657 | hycP | - | 100% (220) | hydrogenase HycP |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0088 | hycP | 1e-81 | 69.09% (220) | hydrogenase HycP |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0085 | hycP | 1e-81 | 69.09% (220) | hydrogenase HycP |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_5111 | - | 3e-84 | 73.18% (220) | hydrogenase 4 membrane component |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1894 | hycP | 1e-116 | 99.09% (220) | hydrogenase HycP |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0088|M.bovis_AF2122/97 MSNANFSILVDFAAGGLVLASVLIVWRRDLRAIVRLLAWQGAALAAIPLL
Rv0085|M.tuberculosis_H37Rv MSNANFSILVDFAAGGLVLASVLIVWRRDLRAIVRLLAWQGAALAAIPLL
MAV_5111|M.avium_104 MSDADFAILIDFAAGGLVLAAVLIVWRRDLRAIVGLLAWQGVALAAIPLV
MMAR_1657|M.marinum_M MTDLNYAGVLDLAPGGLVLTAVLIVWRRDLRSIVWLLAAQGLALAAIPMI
MUL_1894|M.ulcerans_Agy99 MTDLNYAGVLDLAPAGLVLTAVLIVWRRDLRSIVWLLAAQGLALAAIPMI
*:: ::: ::*:*..****::**********:** *** ** ******::
Mb0088|M.bovis_AF2122/97 RGIRDNDRALIAVGIAVLALRALVLPWLLARAVGAEAAAQREATPLVNTA
Rv0085|M.tuberculosis_H37Rv RGIRDNDRALIAVGIAVLALRALVLPWLLARAVGAEAAAQREATPLVNTA
MAV_5111|M.avium_104 RGVHEADAALIVVGGAVLVLRAVVLPRLLARAVGAEQRDQRDATPLVNTA
MMAR_1657|M.marinum_M LGVHSHDWELVGIGLALFALRAVVLPWLLARALGAEKHEQREATPLVNTT
MUL_1894|M.ulcerans_Agy99 LGVHSHDWELVGIGLALFALRAVVLPWLLARALGAEKHEQREATPLVNTT
*::. * *: :* *::.***:*** *****:*** **:*******:
Mb0088|M.bovis_AF2122/97 SSLLITAGLTLTAFAITQPVVNLEPGVTINAVPAAFAVVLIALFVMTTRL
Rv0085|M.tuberculosis_H37Rv SSLLITAGLTLTAFAITQPVVNLEPGVTINAVPAAFAVVLIALFVMTTRL
MAV_5111|M.avium_104 TSLLIAAALTVVAFAVSQPIANLEPSATTNAVPAAFGVVLIALFVMTTRR
MMAR_1657|M.marinum_M ASLLIAAVLTVVALAVTRPIVDLDSSAAVSAVPGGLAVVLIALFVMVTRR
MUL_1894|M.ulcerans_Agy99 ASLLIAAVLTVVALAVTRPIVDLDSSAAVSAVPGGLAVVLIALFVMVTRR
:****:* **:.*:*:::*:.:*:...: .***..:.*********.**
Mb0088|M.bovis_AF2122/97 HAVSQAAGFLMLDNGIAATAFLLTAGVPLIVELGASLDVLFAVIVIGVLT
Rv0085|M.tuberculosis_H37Rv HAVSQAAGFLMLDNGIAATAFLLTAGVPLIVELGASLDVLFAVIVIGVLT
MAV_5111|M.avium_104 HAVSQAAGFLMLDNGIAATAFLLTAGVPLIVELGASLDVLFAVIVIGVLT
MMAR_1657|M.marinum_M HAISQAAGFLMLDNGIAATAFLLTAGVPLIVEMGASLDVLFALIVLGVLT
MUL_1894|M.ulcerans_Agy99 HAISQAAGFLMLDNGIAATAFLLTAGVPLIVEMGASLDVLFALIVLGVLT
**:*****************************:*********:**:****
Mb0088|M.bovis_AF2122/97 GRLRRIFGDADLDKLRELRD
Rv0085|M.tuberculosis_H37Rv GRLRRIFGDADLDKLRELRD
MAV_5111|M.avium_104 GRLRRAFGGADLDQLQELRD
MMAR_1657|M.marinum_M GRLRRTFGGTDLDRLQELRD
MUL_1894|M.ulcerans_Agy99 GRLRRTFGGTDLDRLQEPRD
***** **.:***:*:* **