For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRIVGWAATVAVLAALNPLAPPAKAEPTCGVNLNAPEIQAAIKSLPTLPQSYPWDTNPRSFDPSSNYNPC STLSAVIISVEGATGSSPDLVLLFHNGTSAGIATTKAYSFTTLNAAKTTDDTVALDYKDGRDVCTACPGP ITTVRYEWKGDHVAMLDPAPAW
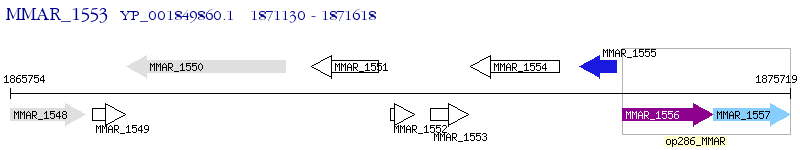
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1553 | - | - | 100% (162) | hypothetical protein MMAR_1553 |
| M. marinum M | MMAR_4965 | - | 8e-27 | 43.67% (158) | hypothetical protein MMAR_4965 |
| M. marinum M | MMAR_4190 | lprE | 1e-15 | 33.33% (144) | lipoprotein LprE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3513c | - | 3e-26 | 41.03% (156) | hypothetical protein Mb3513c |
| M. gilvum PYR-GCK | Mflv_2224 | - | 6e-18 | 35.42% (144) | LprE |
| M. tuberculosis H37Rv | Rv3483c | - | 3e-26 | 41.03% (156) | hypothetical protein Rv3483c |
| M. leprae Br4923 | MLBr_01099 | - | 3e-16 | 35.46% (141) | putative lipoprotein |
| M. abscessus ATCC 19977 | MAB_1402c | - | 2e-12 | 28.57% (147) | putative lipoprotein LprE precursor |
| M. avium 104 | MAV_1400 | - | 3e-16 | 35.17% (145) | LprE protein |
| M. smegmatis MC2 155 | MSMEG_5043 | - | 3e-15 | 35.77% (137) | LprE protein |
| M. thermoresistible (build 8) | TH_2232 | lprE | 8e-15 | 34.06% (138) | PROBABLE LIPOPROTEIN LPRE |
| M. ulcerans Agy99 | MUL_3477 | - | 8e-92 | 96.91% (162) | hypothetical protein MUL_3477 |
| M. vanbaalenii PYR-1 | Mvan_4471 | - | 3e-17 | 34.03% (144) | LprE |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5043|M.smegmatis_MC2_155 -------------------------MKKV--AGVVLATLALAGCGGGDST
TH_2232|M.thermoresistible__bu -------------------------MSRLGIGSVALAVTLLSGCGAGDST
MLBr_01099|M.leprae_Br4923 --------------MRDVWSLPCR-KSLLGVAAVVLVSGTLTGCSSGDST
MAV_1400|M.avium_104 -----------------MWSPPCRTEPLTGSAAVALIAVTLTGCGSGDST
Mflv_2224|M.gilvum_PYR-GCK ---------------MPSLTVRCPTVHWPATVLSVAAALTVAACSPGDST
Mvan_4471|M.vanbaalenii_PYR-1 -------------------MKRAATIG---CVLG-AAALVATACSPGDST
MAB_1402c|M.abscessus_ATCC_199 --------------------MVLGTTPPVLTTLLVVLSALACACTGGDST
MMAR_1553|M.marinum_M ---------------------------------------MRIVGWAATVA
MUL_3477|M.ulcerans_Agy99 ---------------------------------------MRIAGWAATVA
Mb3513c|M.bovis_AF2122/97 MSDEIDPDWPAPAYQPSDDVDTTPPAPGGSWPTAWLVALVVLACVAAAVV
Rv3483c|M.tuberculosis_H37Rv MSDEIDPDWPAPAYQPSDDVDTTPPAPGGSWPTAWLVALVVLACVAAAVV
. .
MSMEG_5043|M.smegmatis_MC2_155 VSKTPGPTASQPATAAPETTVAAP----APTS-----AEPDPCTVNLAAP
TH_2232|M.thermoresistible__bu VSLTPEPTAPSPGAAAPDTPSAAPPVLETPAPG----AQADPCAVNLVAP
MLBr_01099|M.leprae_Br4923 VAKTPVPPSTTTGTISTIISSAPSPPFATAAPPTSNTPPDDPCAVNLASP
MAV_1400|M.avium_104 VAKT--PQARTTSETPSIT--APATPSAAAAQPGVGQP-ADPCAVNLAAP
Mflv_2224|M.gilvum_PYR-GCK VSKTPEQTAPAAAPPAASPPPQSTGPLPGPAP------ADDPCAIDLAAP
Mvan_4471|M.vanbaalenii_PYR-1 VSKTPDQRTPAAAASASAQPPAPAGPLPGPAP------VDDPCDIDLAAP
MAB_1402c|M.abscessus_ATCC_199 VSKTPTNTIASPSSTPPSPGQPSAPVKPKLPS------TKDDCAVNLKDP
MMAR_1553|M.marinum_M VLAALN----------PLAPPAKAEPTCGVNLN---------------AP
MUL_3477|M.ulcerans_Agy99 VLAALN----------PLAPPAQAEPTCGVNLN---------------VP
Mb3513c|M.bovis_AF2122/97 AYAGMHRVRPGANQAAPATTSAPARPTSPASQVG--------PCGPDEAT
Rv3483c|M.tuberculosis_H37Rv AYAGMHRVRPGANQAAPATTSAPARPTSPASQVG--------PCGPDEAT
. . . .
MSMEG_5043|M.smegmatis_MC2_155 EIARAVSELPRDPRSNQAWSPEPI----AGNYNECAQLSAVIVRANTNAE
TH_2232|M.thermoresistible__bu EVLRAIAELPPDPRSGQAWSSEPI----AGNYNECATLSAVIVRANTNAE
MLBr_01099|M.leprae_Br4923 TIARVVSELPRDPRSAQPWNPEPL----AGNYNECAQLSAVIIKANTNAV
MAV_1400|M.avium_104 TIVKVVSELPRDPRSQQPWNPEPL----AGNYNQCAQLSAVIVKANTNAI
Mflv_2224|M.gilvum_PYR-GCK EIAQAVSELPRDPRSGQGWNPEPL----AGNYSECAQLSAVIIKANTNQP
Mvan_4471|M.vanbaalenii_PYR-1 EIARAVSELPRDPRSNQGWNPEPL----AGNYNECAQLSAVIVKANTNSE
MAB_1402c|M.abscessus_ATCC_199 AVASAIALLPPAPNNEATWNPVPV----AGNYNKCAPLSAIIVAADTHEP
MMAR_1553|M.marinum_M EIQAAIKSLPTLPQS-YPWDTNPRSFDPSSNYNPCSTLSAVIISVEGATG
MUL_3477|M.ulcerans_Agy99 EIQAAIKSLPTLPQS-YPWDTNPRSFDPSSNYNPCSTLSAVIIRVEGATG
Mb3513c|M.bovis_AF2122/97 AVRAALAQLAPDSKTGRPWNSTPE----DSNYDPCADLSAVLVTVQDATN
Rv3483c|M.tuberculosis_H37Rv AVRAALAQLAPDSKTGRPWNSTPE----DSNYDPCADLSAVLVTVQDATN
: .: *. ... *.. * .**. *: ***::: .:
MSMEG_5043|M.smegmatis_MC2_155 NPNTRAVLFHLGKFIPTGVPDTYGFNDIDKTQSTGDTVALQYSG------
TH_2232|M.thermoresistible__bu NPNTRAVMFHLGKFIPTGVPDTYGFNAIDTSAGSGDTVALRYDS------
MLBr_01099|M.leprae_Br4923 NPTTRAVLFHLGRFIPQGVPDTYGFNGIDPAQTTGDTVALTYPS------
MAV_1400|M.avium_104 SPSTRAVLFHLGQFIPQGVPDTYGFNGIDPAQTTGDTVALTYPG------
Mflv_2224|M.gilvum_PYR-GCK NPNTRAVLFHQGKFIPTGVPDTYGFNGLDTTQTTGDTVALKYSS------
Mvan_4471|M.vanbaalenii_PYR-1 NPNTRAVLFHQGKFIPTGVPDTYGFNGLDSTQTTGDTVALKYSG------
MAB_1402c|M.abscessus_ATCC_199 QPPTRAVFFHLGGVISHGVPDTYGYNAIDLSASTLDTVVLNFSN------
MMAR_1553|M.marinum_M SSPDLVLLFHNGTSAGIATTKAYSFTTLNAAKTTDDTVALDYKDGRDVCT
MUL_3477|M.ulcerans_Agy99 SSPDLVLLFHNGTSAGIATTKAYSFTTLNAAKTTDDTVALDYKDGRDVCT
Mb3513c|M.bovis_AF2122/97 SSPDQALMFHRGTFVGTATPRAYPFTNLIGPASTNDIVVLSYRT-RQSCD
Rv3483c|M.tuberculosis_H37Rv SSPDQALMFHRGTFVGTATPRAYPFTNLIGPASTNDIVVLSYRT-RQSCD
.. .::** * ... :* :. : . : * *.* :
MSMEG_5043|M.smegmatis_MC2_155 -GFHGLPSTVRFRWNGSGVELMGNVG-------
TH_2232|M.thermoresistible__bu -GIEGLSGVVKFRWNGNGVELVGNTG-------
MLBr_01099|M.leprae_Br4923 -SIDGLATAVRFHWNGNAVELISNIAGG-----
MAV_1400|M.avium_104 -GLSGLSTDVKFHWNGNAVELIGNTPAH-----
Mflv_2224|M.gilvum_PYR-GCK -GMPGLDSVVRFRWNGSGVELIGNTPG------
Mvan_4471|M.vanbaalenii_PYR-1 -GVPGLDSIVKFRWNGSGVELIGNTAR------
MAB_1402c|M.abscessus_ATCC_199 -GIPGLESVVSFRWNGTGVEKVQQAGQ------
MMAR_1553|M.marinum_M ACP-GPITTVRYEWKGDHVAMLDPAPAW-----
MUL_3477|M.ulcerans_Agy99 ACP-GPITTVRYEWKGDHVSMLDPAPAW-----
Mb3513c|M.bovis_AF2122/97 GCQDGILTIVGFAWRGDHVQILDSLPELFDAPP
Rv3483c|M.tuberculosis_H37Rv GCQDGILTIVGFAWRGDHVQILDSLPELFDAPP
* * : *.* * :