For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGWEFGVLLILVAVLLAFLAPRFLPRGPRGALASGTLLVTGVSPRPDDAVGEQYVTITGVINGPTVNEYT VYGRMAVDVDQWPSTGQVLPVVYSPKNPDNWNFALEEPPED
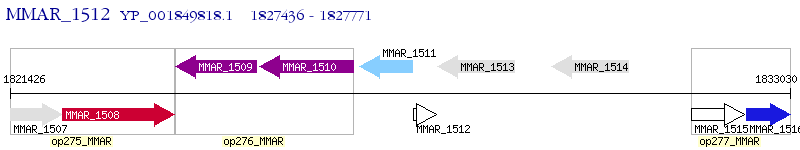
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1512 | - | - | 100% (111) | hypothetical protein MMAR_1512 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4453 | - | 3e-29 | 54.63% (108) | hypothetical protein Mflv_4453 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00663 | - | 7e-39 | 70.48% (105) | hypothetical protein MLBr_00663 |
| M. abscessus ATCC 19977 | MAB_3484c | - | 3e-25 | 50.46% (109) | hypothetical protein MAB_3484c |
| M. avium 104 | MAV_4016 | - | 1e-43 | 74.07% (108) | hypothetical protein MAV_4016 |
| M. smegmatis MC2 155 | MSMEG_2084 | - | 2e-35 | 65.74% (108) | hypothetical protein MSMEG_2084 |
| M. thermoresistible (build 8) | TH_2150 | - | 5e-36 | 63.64% (110) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2429 | - | 1e-62 | 100.00% (111) | hypothetical protein MUL_2429 |
| M. vanbaalenii PYR-1 | Mvan_1909 | - | 3e-32 | 59.26% (108) | hypothetical protein Mvan_1909 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4453|M.gilvum_PYR-GCK -------MFEFAMILLLVGALVLLARPLLARRRGV-GNGWVPGTLLVTGV
Mvan_1909|M.vanbaalenii_PYR-1 MSSYHGPVFEFAMILLLVGALVLLARPFLMRRRGV-GSDWAQGTLLVTGV
TH_2150|M.thermoresistible__bu -------VWELAVLMLLIGAIVVLAAPWIRRRAGIRGDDVAHGTLLVTGV
MMAR_1512|M.marinum_M ------MGWEFGVLLILVAVLLAFLAPRFLP-RGPRG-ALASGTLLVTGV
MUL_2429|M.ulcerans_Agy99 ------MGWEFGVLLILVAVLLAFLAPRFLP-RGPRG-ALASGTLLVTGV
MAV_4016|M.avium_104 -------MWEFGLLVLLVAVLAVFLVQRFVP-RGPRG-ELLSGTLLVTGV
MLBr_00663|M.leprae_Br4923 -------MWEFGLLFLLVAILGVFLAKRFLP-CES---DLASGTLLVTGV
MSMEG_2084|M.smegmatis_MC2_155 -------MWEFGILLLLVGALVLLLAPRIMRRRGMGG-DLTQGTLLVTGV
MAB_3484c|M.abscessus_ATCC_199 -------MWEFLVLLLIGGSLALLIMQMRRGRQAQPGMNAVQGTLLVTGV
:*: ::.:: . : : ********
Mflv_4453|M.gilvum_PYR-GCK SPRPDGVSGEQFVTITGVINGPTVNEYTVYTRMTSDVNSWPTMGQLIDVM
Mvan_1909|M.vanbaalenii_PYR-1 SPRPDGVSGEQFVTITGVINGPTVNEYTVYTRMTVDVDDWPTMGQLIPVM
TH_2150|M.thermoresistible__bu SPRPDGVTGEQFVTITGVINGPTVNEHVVYQRMAVDVEQWPTMGQLIPVV
MMAR_1512|M.marinum_M SPRPDDAVGEQYVTITGVINGPTVNEYTVYGRMAVDVDQWPSTGQVLPVV
MUL_2429|M.ulcerans_Agy99 SPRPDDAVGEQYVTITGVINGPTVNEYTVYGRMAVDVDQWPSTGQVLPVV
MAV_4016|M.avium_104 SPRPE-ATGEQYVTIAGVINGPTVNEHAVYQRMAVDVNQWPTIGQLFPVL
MLBr_00663|M.leprae_Br4923 SPRPDDAHGEQFVTIAGFISGPTVSEYSVYRRMVVDVGKWPAMGQLHPVV
MSMEG_2084|M.smegmatis_MC2_155 SPRPD-AEGEQFVTITGVISGPTVREHVVYQRMAVDVNDWPSMGALFPVV
MAB_3484c|M.abscessus_ATCC_199 SPRPEGVTGEQLVTITGALNGPTVAEYITYRQIVRDVNDWPRIGDLIPVL
****: . *** ***:* :.**** *: .* ::. ** .** * : *:
Mflv_4453|M.gilvum_PYR-GCK YNPGNPEKWAFGSRPEP------------------LPPTDPPPPAPPAPY
Mvan_1909|M.vanbaalenii_PYR-1 YSPGNPEKWAFGSPPAP------------------PAPTEPPP------Y
TH_2150|M.thermoresistible__bu YSARNPDNWHFAPPETP------------------ESTVSDPQ-------
MMAR_1512|M.marinum_M YSPKNPDNWNFALEEPP------------------ED-------------
MUL_2429|M.ulcerans_Agy99 YSPKNPDNWNFALEEPP------------------ED-------------
MAV_4016|M.avium_104 YSSKNPDNWRFAPPAPP------------------EPSPPSQPGG-----
MLBr_00663|M.leprae_Br4923 YSPKNPDNWKFALPD-----------------------------------
MSMEG_2084|M.smegmatis_MC2_155 YSPSNPDKWAFAPTDTP------------------PPSSPPPPPPRPYEP
MAB_3484c|M.abscessus_ATCC_199 YPPKNPDRWNILLAPLPGPTPPQYPSGPPPMENIDPSPPPPPPPPGPVEG
* . **:.* :
Mflv_4453|M.gilvum_PYR-GCK N----------------------
Mvan_1909|M.vanbaalenii_PYR-1 S----------------------
TH_2150|M.thermoresistible__bu -----------------------
MMAR_1512|M.marinum_M -----------------------
MUL_2429|M.ulcerans_Agy99 -----------------------
MAV_4016|M.avium_104 -----------------------
MLBr_00663|M.leprae_Br4923 -----------------------
MSMEG_2084|M.smegmatis_MC2_155 PPV--------------------
MAB_3484c|M.abscessus_ATCC_199 PPSTGDPAKKHEPPPRYYEPPPL