For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTEIRVLAVANQKGGVAKTTTVASLGAAMVDKGRRVLLVDLDPQGCLTFSLGHDPDKLTVSVHEVLLGEV EPSAALVTTMEGMTLLPANIDLAGAEAMLLMRAGREYALKRALAKLSDEFDVVIIDCPPSLGVLTLNGLT AADEVIVPLQCETLAHRGVGQFLRTVSDVQQITNPDLRLLGALPTLYDPRTTHTRDVLLDVADRYSLAVL APPIPRTVRFAEATASGSSVLAGRKNKGALAYRELAQALLKHWKTGKPLPTFTVEP
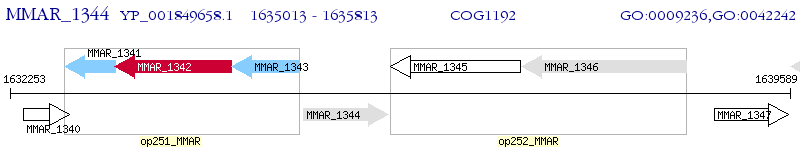
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1344 | - | - | 100% (266) | Soj/ParA-related protein |
| M. marinum M | MMAR_2523 | - | 3e-52 | 44.44% (252) | Soj family ATPase |
| M. marinum M | MMAR_5482 | parA | 3e-46 | 43.65% (252) | chromosome partitioning protein ParA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3239c | - | 1e-136 | 91.70% (265) | SOJ/PARA-like protein |
| M. gilvum PYR-GCK | Mflv_4678 | - | 1e-125 | 85.50% (262) | cobyrinic acid a,c-diamide synthase |
| M. tuberculosis H37Rv | Rv3213c | - | 1e-136 | 91.32% (265) | SOJ/PARA-like protein |
| M. leprae Br4923 | MLBr_01367 | - | 3e-51 | 43.95% (248) | putative regulatory protein |
| M. abscessus ATCC 19977 | MAB_3533c | - | 1e-123 | 84.56% (259) | putative cobyrinic acid a,c-diamide synthase |
| M. avium 104 | MAV_4161 | - | 1e-136 | 91.70% (265) | cobyrinic acid a,c-diamide synthase |
| M. smegmatis MC2 155 | MSMEG_1927 | - | 1e-129 | 88.12% (261) | cobyrinic acid a,c-diamide synthase |
| M. thermoresistible (build 8) | TH_3319 | - | 1e-129 | 88.51% (261) | PUTATIVE cobyrinic Acid a,c-diamide synthase |
| M. ulcerans Agy99 | MUL_2535 | - | 1e-147 | 99.25% (266) | Soj/ParA-related protein |
| M. vanbaalenii PYR-1 | Mvan_1789 | - | 1e-128 | 87.74% (261) | cobyrinic acid a,c-diamide synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1344|M.marinum_M ----------------------------------MTEIRVLAVANQKGGV
MUL_2535|M.ulcerans_Agy99 ----------------------------------MTEIRVLAVANQKGGV
Mb3239c|M.bovis_AF2122/97 ----------------------------------MTDTRVLAVANQKGGV
Rv3213c|M.tuberculosis_H37Rv ----------------------------------MTDTRVLAVANQKGGV
MAV_4161|M.avium_104 ----------------------------------MSKTRVLAVANQKGGV
MSMEG_1927|M.smegmatis_MC2_155 ------------------------------------MTRVLAVANQKGGV
TH_3319|M.thermoresistible__bu VPEQHQPVLPPLLITHDTRFCRTSGDHSRGRVRMGAVTRVLAVANQKGGV
Mflv_4678|M.gilvum_PYR-GCK ---------------------------------MGTVTRVLAVANQKGGV
Mvan_1789|M.vanbaalenii_PYR-1 ------------------------------------MTRVLAVANQKGGV
MAB_3533c|M.abscessus_ATCC_199 ------------------------------------MTRVLAVANQKGGV
MLBr_01367|M.leprae_Br4923 -----MIDHLDSVVEIGLTGRPPRAIPEPRPRSSHGPAKVVAMCNQKGGV
:*:*:.******
MMAR_1344|M.marinum_M AKTTTVASLGAAMVDKGRRVLLVDLDPQGCLTFSLGHDPDKLTVSVHEVL
MUL_2535|M.ulcerans_Agy99 AKTTTVASLGAAMVDKGRRVLLVDLDPQGCLTFSLGHDPDKLTVSVHEVL
Mb3239c|M.bovis_AF2122/97 AKTTTVASLGAAMVEKGRRVLLVDLDPQGCLTFSLGQDPDKLPVSVHEVL
Rv3213c|M.tuberculosis_H37Rv AKTTTVASLGAAMVEKGRRVLLVDLDPQGCLTFSLGQDPDKLPVSVHEVL
MAV_4161|M.avium_104 AKTTTVASLGAAMVDEGKRVLLVDLDPQGCLTFSLGHDPDKLPVSVHEVL
MSMEG_1927|M.smegmatis_MC2_155 AKTTTVASIGAALTEQGRRVLLVDLDPQGCLTFSLGHDPDKLPVSVHEVL
TH_3319|M.thermoresistible__bu AKTTTVASLGAAFAEKGIRVLLVDLDPQGCLTFSLGHDPDKLAVSVHEVL
Mflv_4678|M.gilvum_PYR-GCK AKTTTVASVGAAMVEQGKKVLLVDLDPQGSLTFSLGHDPDKLPVSVHEVL
Mvan_1789|M.vanbaalenii_PYR-1 AKTTTVASLGAAMVELGKKVLLVDLDPQGCLTFSLGQDPDKLPVSVHEVL
MAB_3533c|M.abscessus_ATCC_199 AKTTTVASLGAALAQAGKKVLLVDLDPQGCLTFSLGQDPDRLGISVHEVL
MLBr_01367|M.leprae_Br4923 GKTTSTINLGAALTEFGRRVLLVDIDPQGALSAGLGVPHYELDRTIHNLM
.***:. .:***:.: * :*****:****.*: .** .* ::*:::
MMAR_1344|M.marinum_M LG-EVEPSAALVTT-MEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
MUL_2535|M.ulcerans_Agy99 LG-EVEPSAALVTT-LEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
Mb3239c|M.bovis_AF2122/97 LG-EVEPNAVLVTT-MEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
Rv3213c|M.tuberculosis_H37Rv LG-EVEPNAVLVTT-MEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
MAV_4161|M.avium_104 LG-EVEPNAALVDT-AEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
MSMEG_1927|M.smegmatis_MC2_155 LG-DVEPSAALVRT-DEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
TH_3319|M.thermoresistible__bu LG-EVEPGAALVET-AEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
Mflv_4678|M.gilvum_PYR-GCK LG-DVEPDVAIVET-PEGMSLLPANIDLAGAEAMLLMRAGREHALKRALD
Mvan_1789|M.vanbaalenii_PYR-1 LG-DVEPEAALVET-SEGMTLLPANIDLAGAEAMLLMRAGREYALKRALA
MAB_3533c|M.abscessus_ATCC_199 LG-QADIKEALLTT-TEDLTLLPANIDLAGAEAMLLMRAGREYALKRALE
MLBr_01367|M.leprae_Br4923 VEPLVSIDDVLIHTRVRYLDLVPSNIDLSAAEIQLVNEVGREQTLARALH
: .. .:: * . : *:*:****:.** *: ..*** :* ***
MMAR_1344|M.marinum_M KLSDEFDVVIIDCPPSLGVLTLNGLTAADEVIVPLQCETLAHRGVGQFLR
MUL_2535|M.ulcerans_Agy99 KLSDEFDVVIIDCPPSLGVLTLNGLTAADEVIVPLQCETLAHRGVGQFLR
Mb3239c|M.bovis_AF2122/97 KFSDRFDVVIIDCPPSLGVLTLNGLTAADEAIVPLQCEMLAHRGVGQFLR
Rv3213c|M.tuberculosis_H37Rv KFSDRFDVVIIDCPPSLGVLTLNGLTAADKAIVPLQCEMLAHRGVGQFLR
MAV_4161|M.avium_104 KLADRFDVVIVDCPPSLGVLTLNGLTAADEVIVPLQCETLAHRGVGQFLR
MSMEG_1927|M.smegmatis_MC2_155 KLDGDFDVVIIDCPPSLGVLTLNGLTAAHDVIVPLQCETLAHRGVGQFLR
TH_3319|M.thermoresistible__bu KVSDDFEVIIIDCPPSLGVLTLNGLTAAQEAIVPLQCETLAHRGVGQFLR
Mflv_4678|M.gilvum_PYR-GCK KVSAEFDVVIIDCPPSLGVLTLNGLTAADEVVVPLQCETLAHRGVGQFLR
Mvan_1789|M.vanbaalenii_PYR-1 KISDAFDVILIDCPPSLGVLTLNGLTAADDVIVPLQCETLAHRGVGQFLR
MAB_3533c|M.abscessus_ATCC_199 SLGDQFDVVIIDCPPSLGVLTLNGLTAADSVMVPLQCETLAHRGVGQFLR
MLBr_01367|M.leprae_Br4923 PVLDRYDYVLIDCQPSLGLLTVNGLACAEGVVIPTECEFFSLRGLALLTD
. :: :::** ****:**:***:.*. .::* :** :: **:. :
MMAR_1344|M.marinum_M TVSDVQQITNPDLRLLGALPTLYDPRTTHTRDVLLDVADRYSLAVLAPPI
MUL_2535|M.ulcerans_Agy99 TVSDVQQITNPDLRLLGALPTLYDPRTTHTRDVLLDVADRYSLAVLAPPI
Mb3239c|M.bovis_AF2122/97 TVADVQQITNPNLRLLGALPTLYDSRTTHTRDVLLDVADRYDLQVLAPPI
Rv3213c|M.tuberculosis_H37Rv TVADVQQITNPNLRLLGALPTLYDSRTTHTRDVLLDVADRYDLQVLAPPI
MAV_4161|M.avium_104 TVTDVQQITNPDLRLLGALPTLYDSRTTHTRDVLVDVADRYSLPVLAPPI
MSMEG_1927|M.smegmatis_MC2_155 TISDVQQITNPDLKLLGALPTLYDSRTTHSRDVLLDVADRYELPVLAPPI
TH_3319|M.thermoresistible__bu TIDDVQQITNPDLTLLGALPTLYDSRTTHSRDVLLDVADRYGLPVLAPPI
Mflv_4678|M.gilvum_PYR-GCK TVSDVQAITNPDLKMLGALPTLYDSRTTHSRDVLFDVVDRYGLPVLAPPI
Mvan_1789|M.vanbaalenii_PYR-1 TINDVQQITNADLKLLGALPTLYDSRTTHSRDVLFDVVDRYNLPVLAPPI
MAB_3533c|M.abscessus_ATCC_199 TVTDVQQITNPRLTLLGALPTLYDSRTTHSRDVLLDVADRYQLPVLAPPI
MLBr_01367|M.leprae_Br4923 TVDKVRDRLNPKLEISGILITRYDPRTVNAREVMARVVERFGDLVFDTVI
*: .*: *. * : * * * **.**.::*:*: *.:*: *: . *
MMAR_1344|M.marinum_M PRTVRFAEATASGSSVLAGR-KNKGALAYRELAQALLKHWKTGKPLPTFT
MUL_2535|M.ulcerans_Agy99 PRTVRFAEATASGSSVLAGR-KNKGALAYRELAQALLKHWATGKPLPTFT
Mb3239c|M.bovis_AF2122/97 PRTVRFAEASASGSSVMAGR-KNKGAVAYRELAQALLKHWKTGRPLPTFT
Rv3213c|M.tuberculosis_H37Rv PRTVRFAEASASGSSVMAGR-KNKGAVAYRELAQALLKHWKTGRPLPTFT
MAV_4161|M.avium_104 PRTVRFAEASASGSSVMAGR-KNKGAAAYGELAAALLKHWKSGKPLPTFT
MSMEG_1927|M.smegmatis_MC2_155 PRTVRFAEASASGSSVLAGR-KSKGAIAYREFADALLRHWKSGRKMPTFT
TH_3319|M.thermoresistible__bu PRTVRFAEASASGASVLAGR-KNKGALAYRELAEALLKHWETGEEMATFT
Mflv_4678|M.gilvum_PYR-GCK PRTVRFAEASASGASVLTGR-KNKGAMAYREFADALLKHWKTGKDLATFT
Mvan_1789|M.vanbaalenii_PYR-1 PRTVRFAEATASGCSVLAGR-KNKGAMAYREFAAALLKHWKNGKPLETFT
MAB_3533c|M.abscessus_ATCC_199 PRTVRFAEASASGASVLAGR-KNKGALAYRELAQSLVKHWKSGKAMPTFT
MLBr_01367|M.leprae_Br4923 TRTVRFPETSVAGEPITTWAPKSGGARAYRALACEFIDRFGA--------
.*****.*::.:* .: : *. ** ** :* :: ::
MMAR_1344|M.marinum_M VEP-
MUL_2535|M.ulcerans_Agy99 VEP-
Mb3239c|M.bovis_AF2122/97 VDL-
Rv3213c|M.tuberculosis_H37Rv VDL-
MAV_4161|M.avium_104 VEV-
MSMEG_1927|M.smegmatis_MC2_155 PEVV
TH_3319|M.thermoresistible__bu PEL-
Mflv_4678|M.gilvum_PYR-GCK PDL-
Mvan_1789|M.vanbaalenii_PYR-1 PEV-
MAB_3533c|M.abscessus_ATCC_199 PPVD
MLBr_01367|M.leprae_Br4923 ----