For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTTTTEPAKPGTLDRLRARYGWFDHAMRAAKRFNEQRGGFFAGGLTYYTIFALFPLLMVGFAVFGFVLAS HPDLLKTIDGKIRESVSGALSQQLVGLMNSAIDARASVGLIGLATASWAGLTWMSHLRVALSEMWAHPVQ RVGYVRTKLSDLTAMIGTFLVTMLTIALTALGHLRPMATVLSWFGIPELAIFNVVFRLISILISVLVAWL MFTWMIARLPRQPGRLGTSMQAGLMAAIGFELFKQVASIYLRTVLRSPAGAAFGPVLGLMVFANVTAYLV LFATAWAATKDPYSEHIEPPGPAIISPRIQVEEGLGVRQTVAALIAGAVGALTISRLFRRRR
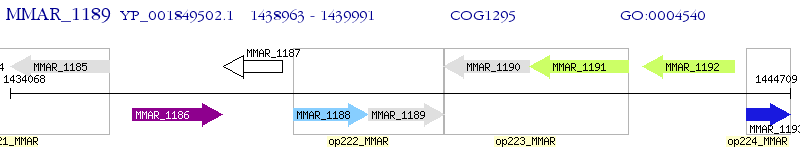
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1189 | - | - | 100% (342) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3368c | - | 1e-115 | 70.07% (284) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4870 | - | 1e-100 | 54.07% (344) | putative ribonuclease |
| M. tuberculosis H37Rv | Rv3335c | - | 1e-115 | 70.07% (284) | integral membrane protein |
| M. leprae Br4923 | MLBr_00687 | - | 1e-100 | 61.46% (288) | hypothetical protein MLBr_00687 |
| M. abscessus ATCC 19977 | MAB_3682c | - | 6e-97 | 50.89% (336) | hypothetical protein MAB_3682c |
| M. avium 104 | MAV_2739 | - | 1e-11 | 26.80% (250) | hypothetical protein MAV_2739 |
| M. smegmatis MC2 155 | MSMEG_1658 | - | 1e-107 | 57.91% (335) | ribonuclease, putative |
| M. thermoresistible (build 8) | TH_1975 | - | 1e-105 | 57.57% (337) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1442 | - | 0.0 | 99.42% (342) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1561 | - | 1e-100 | 54.62% (346) | putative ribonuclease |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1189|M.marinum_M MTTTTEPAK----PGTLDRLRARYGWFDHAMRAAKRFNEQRGGFFAGGLT
MUL_1442|M.ulcerans_Agy99 MTTTTEPAK----PGTLDRLRARYGWFDHAMRAAKRFNEQRGGFFAGGLT
Mb3368c|M.bovis_AF2122/97 ---MGELAE----PGVLDRLRARFGWLDHVVRAFTRFNDRNGSLFAAGLT
Rv3335c|M.tuberculosis_H37Rv ---MGELAE----PGVLDRLRARFGWLDHVVRAFTRFNDRNGSLFAAGLT
MLBr_00687|M.leprae_Br4923 ---MTEPAN----TGIIDRLRGRFRWLDHVVRAHQRFDERNGSFFAAGLT
Mflv_4870|M.gilvum_PYR-GCK MTTQSATADPEDKPGFLDRQRARRPWFDHVMRAQERYKDSKGDFYAAGIT
Mvan_1561|M.vanbaalenii_PYR-1 MTAQPAPTDTEDKPGFLDRMRARMPWFDHVMRAQERYKDAKGDFYAAGIT
MSMEG_1658|M.smegmatis_MC2_155 ---MTEPADESAKPGLLDRLKARWPWFDHVMRAQERYNDSKGDFYAAGIT
TH_1975|M.thermoresistible__bu ---VTEPSE----PGVLQRLRARYAWFDHVLRAQERYTEARGNHFAAGIT
MAB_3682c|M.abscessus_ATCC_199 --MVAEDDK----PGILDRYRARFPWFDHIMRMQERYGKVNGNFFAAGIT
MAV_2739|M.avium_104 ----------------------------MIVGVVYKYNDDQGGYLAALIT
: :: . .*. *. :*
MMAR_1189|M.marinum_M YYTIFALFPLLMVGFAVFGFVLASHPDLLKTIDGKIRESVSGALSQQLVG
MUL_1442|M.ulcerans_Agy99 YYTIFALFPLLMVGFAVFGFVLASHPDLLKTINGKIRESVSGALSQQLVG
Mb3368c|M.bovis_AF2122/97 YYTIFAIFPLLMVGFGVGGFALSRRPELLTTLEERIRTSVSGAVGQQLVD
Rv3335c|M.tuberculosis_H37Rv YYTIFAIFPLLMVGFGVGGFALSRRPELLTTLEERIRTSVSGAVGQQLVD
MLBr_00687|M.leprae_Br4923 YYTIFALFPMLMVSFSVVGLVLFRRPELLTTIDDHIRSAVSSQLGQELVD
Mflv_4870|M.gilvum_PYR-GCK YFTIFALFPLLMLGFSVAGFVLANQPELLQDVEERIRSTFSGDLGQQLVT
Mvan_1561|M.vanbaalenii_PYR-1 YFTIFALFPLLMLGFSVGGFVLASQPELLATVVERIRSAVSGELGRQLIE
MSMEG_1658|M.smegmatis_MC2_155 YFTIFALFPLLMVGFAVGGFVLASQPELMGQLEDKIRQAVSGDLGAQLVQ
TH_1975|M.thermoresistible__bu YFTVFALFPLLMVGFSIGGFVLAGRPDLLRQIEDRIRTTVSGDASQQLIE
MAB_3682c|M.abscessus_ATCC_199 YFTVFALFPLMMVAFAVMGFVLSRRPDTIAELRGRISGAISGEVGNQLIN
MAV_2739|M.avium_104 YYALVSLFPLLLLLTTGLGVILAGRPDLQAQVLHSTLSEFP-VIGSQLHH
*:::.::**:::: *. * :*: : .. . :*
MMAR_1189|M.marinum_M LMNSAIDARASVGLIGLATASWAGLTWMSHLRVALSEMW-AHPVQRVGYV
MUL_1442|M.ulcerans_Agy99 LMNSAIDARASVGLIGLATASWAGLTWMSHLRVALSEMW-AHPVQRVGYV
Mb3368c|M.bovis_AF2122/97 LMNSAIDARASVGVIGLATAAWVGLGWMWHLREALSQMW-AHPVAPAGYL
Rv3335c|M.tuberculosis_H37Rv LMNSAIDARASVGVIGLATAAWVGLGWMWHLREALSQMW-AHPVAPAGYL
MLBr_00687|M.leprae_Br4923 LMNRAIDARASVGVVGLATAAWAGLCWMSHFRAALTKMWWEHHIDPSSLL
Mflv_4870|M.gilvum_PYR-GCK LMDSAIESRGAVGLIGLATAAWAGLGWMANLREALSQMWGLFRGETPGFV
Mvan_1561|M.vanbaalenii_PYR-1 LMDSAIDSRGTVGVIGLATAAWAGLGWMANLREALSQMWGLYREESPGFV
MSMEG_1658|M.smegmatis_MC2_155 LMDSAIDSRTSVGVIGLATAAWAGLGWMANLREALSQMWGLFREEKPGFV
TH_1975|M.thermoresistible__bu LMDSAIASRTSVGLIGLATATWAGLSWMANLREALSEMWEQ-RPEKGNFV
MAB_3682c|M.abscessus_ATCC_199 LMDTAIASRTSLGIAGLATAAWAGLGWMANVRAALSAMWEQ-PNDSENFV
MAV_2739|M.avium_104 PEG--LSGGAAGVIVGLAGALYGGLGVGQAVQNAMDSVWAVPKHKRPNPI
. : . : : *** * : ** .: *: :* . :
MMAR_1189|M.marinum_M RTKLSDLTAMIGTFLVTMLTIALTALGHLRPMATVLSWFGIPELAIFNVV
MUL_1442|M.ulcerans_Agy99 RTKLSDLTAMIGTFLVTMLTIALTALGHLRPMATVLSWFGIPELAIFNVV
Mb3368c|M.bovis_AF2122/97 RTKLSDLAAMVGTFVVIVATIALTVLGHARPMAAVLRWLEIPQFSVFDEI
Rv3335c|M.tuberculosis_H37Rv RTKLSDLAAMVGTFVVIVATIALTVLGHARPMAAVLRWLEIPQFSVFDEI
MLBr_00687|M.leprae_Br4923 RNKLSDLLAMVGTFVVFLTTLALTVLSHQAPMAAILRCIGITHFFVFGVI
Mflv_4870|M.gilvum_PYR-GCK KTKLSDLVALLTTFVAIIVTLALSALGNSSLVENILTRIGFPDSPVLTVV
Mvan_1561|M.vanbaalenii_PYR-1 RTKLSDLLALLSAFAAIVVTLALTALGNTSVMAGVLARLGIEDAPGLSLM
MSMEG_1658|M.smegmatis_MC2_155 RTKLSDLLALLSAFVAIILTIGLTALGSSTAIENVLNRIGMPDSVGLSIL
TH_1975|M.thermoresistible__bu RTKLSDLVALLSLFLALLVTFGLTALGDRAVMRGVLRRLEISEFPGLGIG
MAB_3682c|M.abscessus_ATCC_199 KGKLSDLLALGSTFVALALTIAVTAVGDEKTILRILGWMGLHDVAIPSVL
MAV_2739|M.avium_104 RSRVRSLLLLLVLGSAAIAATLLAAAGQATAGLGFLGKAGLA--------
: :: .* : . : ::. . .* :
MMAR_1189|M.marinum_M FRLISILISVLVAWLMFTWMIARLPRQPGRLGTSMQAGLMAAIGFELFKQ
MUL_1442|M.ulcerans_Agy99 FRLISILISVLVAWLMFTWMIARLPRQPGRLGTSMQAGLMAAIGFELFKQ
Mb3368c|M.bovis_AF2122/97 FRGISVLVSVLVSWVLFTWMIGRLPREPVGLVTAARAGLMAAVGFELFKQ
Rv3335c|M.tuberculosis_H37Rv FRGISVLVSVLVSWVLFTWMIGRLPREPVGLVTAARAGLMAAVGFELFKQ
MLBr_00687|M.leprae_Br4923 FQGASVLVSTLVSWLLFTWMIAWLPRKSVNLVASMRGGLIAAVGFELFKQ
Mflv_4870|M.gilvum_PYR-GCK LRASSITVSVLVSWLLFTWIIARLPRERVGFRSSVRAGLIAALAFEVFKL
Mvan_1561|M.vanbaalenii_PYR-1 LRLASFAVSVLVSWSLFTWMIARLPRESVSFRSSVRAGMIAAVAFEIFKF
MSMEG_1658|M.smegmatis_MC2_155 LWVLSVLMSLFVSWLLFTWMIARLPRETVTFRSSLRAGLLAAVGFEIFKQ
TH_1975|M.thermoresistible__bu LKVASIAVAMLVAWLLFTWVIARLPRTTVSLRSSVRAGLLAAIGFEIFKQ
MAB_3682c|M.abscessus_ATCC_199 LKVVTIAIATVLSWALFTWMIARLPRERLPFSSSVRAGLIAAIGFELFKQ
MAV_2739|M.avium_104 --LTSVAINALICLVLFRLTTAR----ALSYRQVWPGAVAAALVWQLLQW
:. : .:. :* . ..: **: :::::
MMAR_1189|M.marinum_M VASIYLRTVLR--SPAGAAFGPVLGLMVFANVTAYLVLFATAW-------
MUL_1442|M.ulcerans_Agy99 VASIYLRTVLR--SPAGAAFGPVLGLMVFANVTAYLVLFATAW-------
Mb3368c|M.bovis_AF2122/97 VGAIYLQIVLR--SPAGAVFGPVLGLMVFAFVTAWLILFATAW-------
Rv3335c|M.tuberculosis_H37Rv VGAIYLQIVLR--SPAGAVFGPVLGLMVFAFVTAWLILFATAW-------
MLBr_00687|M.leprae_Br4923 VGSIYLQIVLR--SPAGSVFGPVLGVMVFSYFTAYLVLFAASW-------
Mflv_4870|M.gilvum_PYR-GCK VASIYLRSVLT--GPAGATFGPVLGLMVFAYITSRLILFSTCW-------
Mvan_1561|M.vanbaalenii_PYR-1 FGSVYLRSVVT--GPAGATFGPVLGLMVFAYITSRLILFSAAW-------
MSMEG_1658|M.smegmatis_MC2_155 LASIYLKSVIT--GPAGATFGPVLGLMVFAYITARLVLFATAW-------
TH_1975|M.thermoresistible__bu VGSVYLASVTT--GPAGATFGPVLGLMVFAYITSRLILFATAW-------
MAB_3682c|M.abscessus_ATCC_199 VASIYLSVIMH--GPAGSTFGPVLGIMVFAYITARLLLYATAW-------
MAV_2739|M.avium_104 FGAGYVAHVVKRASATNSVFALVLGALAFLFLASTVLVVCAEVNVVLVER
..: *: : ..:.:.*. *** :.* .:: ::: .:
MMAR_1189|M.marinum_M --------AATKDPYS--EHIEPPGPAIISPRIQVEEGLGVRQTVA----
MUL_1442|M.ulcerans_Agy99 --------AATKDPYS--EHIEPPDPAIISPRIQVEEGLGVRQTVA----
Mb3368c|M.bovis_AF2122/97 --------AATASA------------------------------------
Rv3335c|M.tuberculosis_H37Rv --------AATASA------------------------------------
MLBr_00687|M.leprae_Br4923 --------AATAELCM--KPVS------------------VQLPWS----
Mflv_4870|M.gilvum_PYR-GCK --------AATTPENMRENEIPAPPPAVINNRVIARPGLQIWQGLT----
Mvan_1561|M.vanbaalenii_PYR-1 --------AATTPENMAEELIPAPAPAIINNRIINRPGLQAWQAVA----
MSMEG_1658|M.smegmatis_MC2_155 --------AATAEENLEAARVLPPEPVQITTRVQVREGAGVPAMLT----
TH_1975|M.thermoresistible__bu --------AATSADSLAARPVEPPVGARITTRIQVREGVGVVGTLT----
MAB_3682c|M.abscessus_ATCC_199 --------AATSDENRPLAHVPPPEPVVITTRVETVERPSKLGVIA----
MAV_2739|M.avium_104 LYPRALLGAFSDDADLTSADRRTYTKKVKAERVKAFERVSVRLDDAKRSP
* :
MMAR_1189|M.marinum_M ALIAGAVGALTISRLFRRRR
MUL_1442|M.ulcerans_Agy99 ALIAGAVGALTISRLFRRRR
Mb3368c|M.bovis_AF2122/97 --------------------
Rv3335c|M.tuberculosis_H37Rv --------------------
MLBr_00687|M.leprae_Br4923 YLFPGRDESGS---------
Mflv_4870|M.gilvum_PYR-GCK AAAVGAIGALSLSRRRR---
Mvan_1561|M.vanbaalenii_PYR-1 AATAGALGALSLTRWRRR--
MSMEG_1658|M.smegmatis_MC2_155 AAAAGAVGGFGLSRLRFRQ-
TH_1975|M.thermoresistible__bu AAAIGALGALGLSRWLRH--
MAB_3682c|M.abscessus_ATCC_199 AFGAGVIGALGISWLGRGGD
MAV_2739|M.avium_104 AKTVGRLNIFGDVRRPRTP-