For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRIRLLRAKLHQVRVTHRERDYVGSIAVDVDLMEQVGMLPLEEVEVVNLENGNRWSTYLIPADRGSSRIC PNGGGALLCEPGDRLIIFSYATVRSGALRRRPHVARVLLSDADNRVESLFEQVLEQDDDRLRYSVRGCYG DIPAQPDLLLRAETQLLDGPH*
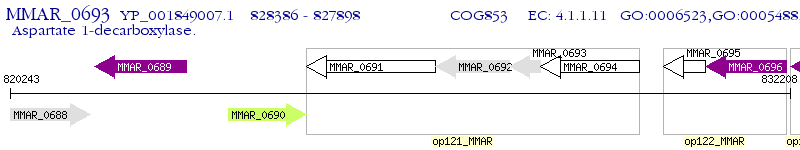
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0693 | panD_1 | - | 100% (162) | aspartate decarboxylase, PanD_1 |
| M. marinum M | MMAR_5104 | panD | 8e-19 | 38.74% (111) | aspartate 1-decarboxylase precursor PanD |
| M. marinum M | MMAR_3177 | panD_2 | 3e-16 | 38.35% (133) | aspartate 1-decarboxylase precursor PanD_2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3631c | panD | 3e-18 | 39.64% (111) | aspartate alpha-decarboxylase |
| M. gilvum PYR-GCK | Mflv_1425 | - | 1e-19 | 41.74% (115) | aspartate alpha-decarboxylase |
| M. tuberculosis H37Rv | Rv3601c | panD | 3e-18 | 39.64% (111) | aspartate alpha-decarboxylase |
| M. leprae Br4923 | MLBr_00231 | panD | 2e-17 | 37.84% (111) | aspartate alpha-decarboxylase |
| M. abscessus ATCC 19977 | MAB_0542 | - | 1e-18 | 39.32% (117) | aspartate 1-decarboxylase precursor |
| M. avium 104 | MAV_0552 | panD | 8e-19 | 38.74% (111) | aspartate alpha-decarboxylase |
| M. smegmatis MC2 155 | MSMEG_0021 | panD | 5e-15 | 38.33% (120) | aspartate 1-decarboxylase |
| M. thermoresistible (build 8) | TH_2350 | panD | 2e-18 | 38.74% (111) | PROBABLE ASPARTATE 1-DECARBOXYLASE PRECURSOR PAND (ASPARTATE |
| M. ulcerans Agy99 | MUL_4183 | panD | 6e-19 | 38.74% (111) | aspartate alpha-decarboxylase |
| M. vanbaalenii PYR-1 | Mvan_5363 | - | 2e-19 | 41.44% (111) | aspartate alpha-decarboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00231|M.leprae_Br4923 -MLRTMLKSKIHRATVTQAYLHYVGSVTIDADLMGAADLLEGEQVTIVDI
MUL_4183|M.ulcerans_Agy99 -----MLKSKIHRATVTQADLHYVGSVTIDADLMDAADLLEGEQVTIVDI
MAV_0552|M.avium_104 -MLRTMLKSKIHRATVTQADLHYVGSVTIDADLMDAADLLEGEQVTIVDI
Mb3631c|M.bovis_AF2122/97 -MLRTMLKSKIHRATVTCADLHYVGSVTIDADLMDAADLLEGEQVTIVDI
Rv3601c|M.tuberculosis_H37Rv -MLRTMLKSKIHRATVTCADLHYVGSVTIDADLMDAADLLEGEQVTIVDI
TH_2350|M.thermoresistible__bu -MFRTMLKSKIHRATVTQADLHYVGSVTIDADLMDAADLLEGEQVTIVDI
Mvan_5363|M.vanbaalenii_PYR-1 -MFRTMLKSKIHRATVTQADLHYVGSVTVDADLMDAADLIEGEQVTIVDI
Mflv_1425|M.gilvum_PYR-GCK -MLRTMLKSKIHRATVTQADLHYVGSVTVDADLMDAADLLEGEQVTIVDV
MAB_0542|M.abscessus_ATCC_1997 MMFRTMMKSKIHRATVTHADLHYVGSVTIDPDLMEAADLLEGEQVTIVDI
MSMEG_0021|M.smegmatis_MC2_155 -MQRVLLASKIHRATVTQADLHYVGSITIDAELMAAADIVEGEQVHVVDI
MMAR_0693|M.marinum_M MSRIRLLRAKLHQVRVTHRERDYVGSIAVDVDLMEQVGMLPLEEVEVVNL
:: :*:*:. ** .****:::* :** ..:: *:* :*::
MLBr_00231|M.leprae_Br4923 NNGARLVTYAIAGERGTGVIGINGAAAHLVHPGDLVILISYGTMEDAEAH
MUL_4183|M.ulcerans_Agy99 DNGARLVTYAITGERGSGVIGINGAAAHLVHPGDLVILIAYGTMQDAEAR
MAV_0552|M.avium_104 DNGARLVTYAITGERGSGVIGINGAAAHLVHPGDLVILIAYGTMEEAEAR
Mb3631c|M.bovis_AF2122/97 DNGARLVTYAITGERGSGVIGINGAAAHLVHPGDLVILIAYATMDDARAR
Rv3601c|M.tuberculosis_H37Rv DNGARLVTYAITGERGSGVIGINGAAAHLVHPGDLVILIAYATMDDARAR
TH_2350|M.thermoresistible__bu TNGARLVTYAITGERGSGVIGINGAAAHLVHPGDLVILIAYGTMSDAEAR
Mvan_5363|M.vanbaalenii_PYR-1 ENGARLVTYVITGERGSGVIGINGAAAHLVHPGDLVILIAYGTMEDAEAR
Mflv_1425|M.gilvum_PYR-GCK DNGARLVTYVITGERGSGVIGINGAAAHLIHPGDLVILIAYGMLDDAAAR
MAB_0542|M.abscessus_ATCC_1997 DNGARLETYAITGTRGSGVIGINGAAAHLVHPGDLVIIIAYGVMTDEEAR
MSMEG_0021|M.smegmatis_MC2_155 TSGARLVTYAITGTPGSGVIGINGAAAHLISPGNLVIIMSFVHLDEAERA
MMAR_0693|M.marinum_M ENGNRWSTYLIPADRGSSRICPNGGGALLCEPGDRLIIFSYATVRSGALR
.* * ** *.. *:. * **..* * **: :*:::: : .
MLBr_00231|M.leprae_Br4923 --AYQPRIVFVDADNKPIDLG---HDPGS----------------VP-LD
MUL_4183|M.ulcerans_Agy99 --AYQPRIVFVDADNKQIDVG---HDPAF----------------VPAFD
MAV_0552|M.avium_104 --AYQPRIVFVDADNKPVDLG---HDPAF----------------VPDFE
Mb3631c|M.bovis_AF2122/97 --TYQPRIVFVDAYNKPIDMG---HDPAF----------------VP---
Rv3601c|M.tuberculosis_H37Rv --TYQPRIVFVDAYNKPIDMG---HDPAF----------------VP---
TH_2350|M.thermoresistible__bu --TYQPRIVFVDADNKPIDLGVHASDPAY----------------VP---
Mvan_5363|M.vanbaalenii_PYR-1 --AYRPRVVFVDADNRPIDLG---VDPAY----------------VP---
Mflv_1425|M.gilvum_PYR-GCK --TYEPRVVFVDADNRVLDLG---ADPAF----------------VP---
MAB_0542|M.abscessus_ATCC_1997 --AFTPRVVFVDSDNKQVELGEHSDDPAY----------------VP---
MSMEG_0021|M.smegmatis_MC2_155 --DHRANVVHVDADNHIVTIG---SDPAQP---------------VP---
MMAR_0693|M.marinum_M RRPHVARVLLSDADNRVESLFEQVLEQDDDRLRYSVRGCYGDIPAQPDLL
. ..:: *: *: : : *
MLBr_00231|M.leprae_Br4923 ISVAAELFDPRIGAR
MUL_4183|M.ulcerans_Agy99 IPGAEELLNPRIGAR
MAV_0552|M.avium_104 IAGAAELLDPRIVAR
Mb3631c|M.bovis_AF2122/97 -ENAGELLDPRLGVG
Rv3601c|M.tuberculosis_H37Rv -ENAGELLDPRLGVG
TH_2350|M.thermoresistible__bu -ADAAELMSPR----
Mvan_5363|M.vanbaalenii_PYR-1 -PDAGELLSPR----
Mflv_1425|M.gilvum_PYR-GCK --DTAELLSPR----
MAB_0542|M.abscessus_ATCC_1997 --ENFGLVSPRGLV-
MSMEG_0021|M.smegmatis_MC2_155 --GAADQLVGRV---
MMAR_0693|M.marinum_M LRAETQLLDGPH---
.