For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGRHELARKRRKPSVLLAAVVAPVAAFFAVAGDIAPSSARDRIDQMAVGDGACCVEIVAATPIELSSKIS SGAVGMGFAAGQFAAASRLRVDRSARLLPVGVAPERGLQVKTILAARSISAVFPEIHEIGGVRPDALRWH PNGLALDVMIPNPGSAEGIALGNEIVAYVLANAERFGMQDAIWRGVYYTPSGATRSRLGHYDHVHVTTTG GGYPTGKEIYLR
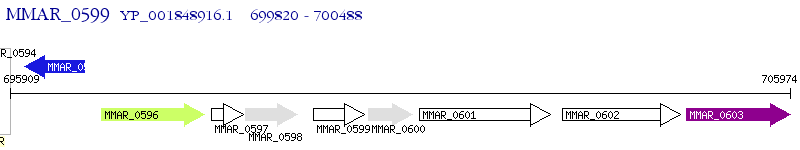
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0599 | - | - | 100% (222) | hypothetical protein MMAR_0599 |
| M. marinum M | MMAR_2569 | - | 2e-42 | 50.28% (179) | hypothetical protein MMAR_2569 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0328 | - | 8e-87 | 68.02% (222) | hypothetical protein Mb0328 |
| M. gilvum PYR-GCK | Mflv_1672 | - | 8e-71 | 59.91% (232) | hypothetical protein Mflv_1672 |
| M. tuberculosis H37Rv | Rv0320 | - | 8e-87 | 68.02% (222) | hypothetical protein Rv0320 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0233 | - | 2e-11 | 32.89% (152) | putative phage tail tape measure protein TMP |
| M. avium 104 | MAV_4829 | - | 2e-61 | 72.73% (154) | hypothetical protein MAV_4829 |
| M. smegmatis MC2 155 | MSMEG_0651 | - | 4e-70 | 60.79% (227) | hypothetical protein MSMEG_0651 |
| M. thermoresistible (build 8) | TH_1403 | - | 3e-46 | 54.65% (172) | NLP/P60 family protein |
| M. ulcerans Agy99 | MUL_0559 | - | 1e-120 | 96.85% (222) | hypothetical protein MUL_0559 |
| M. vanbaalenii PYR-1 | Mvan_5081 | - | 2e-68 | 58.80% (233) | hypothetical protein Mvan_5081 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 MAVELASGYVSLSVRVGATSGLGKLFDNAQKQATSAGKKTGEAFAKALAS
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 EAKAAEEQVKKLTDTVTKSRDKEADQAGKLKVALEKLNEARQGGVSGSKL
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 TALEESHAAALRKQQAAAQDLAKDLDAVTRAQRRATDAQAAVGRAPKNVQ
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 NRVSQLLSGSADAAGHAGGLAGRRFGDSFSGALRSTGIVAAGTAVGNLAA
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 NAMTKAASFATSGVSAVVTKGLDFEKTMNTLSGVTGASADVMQRFRDTAK
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 ALGNDMTLSNTSAADAAQAMTELAKAGFSVDESIAGAKGTLQLAAAAQVD
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 AGKAAEIQANALLAFGLKADYASKAADVLSNAANASSAEITDVAQALQSG
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 GAVANQFGLKLEDTAAAIGLLANNGIKGSDAGTLLKSALLALTDTSNPAQ
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 GAIEELGLTVYDAQGRFVGLEKLFGDLQAASQRMTPEMYQAATTTLFGSD
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 AARLAGIAAKDGAAGYDQMRDAMEKQGSAAKLAAAQNQGLPGVIERLKNA
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 AETLAITLFEKVQGPLSSIGDGLTGFTNKMQDAFENPAVSQAAGAIGAAL
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu ---------------------------------------------VLTLS
MAB_0233|M.abscessus_ATCC_1997 SGIGAAFGRILSAVGPALVGGLSNAVNLIVRFKDFLIPLVAGLVAYKTVM
MMAR_0599|M.marinum_M ------------------MGRHELAR------------------------
MUL_0559|M.ulcerans_Agy99 ------------------MGRHGLAR------------------------
Mb0328|M.bovis_AF2122/97 ------------------MGRHELAR------------------------
Rv0320|M.tuberculosis_H37Rv ------------------MGRHELAR------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------MLLGVGRHSLPK------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------MLLGVGRHSIAK------------------------
MSMEG_0651|M.smegmatis_MC2_155 ---------------MLRVGRHSLASP-----------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu MGQVQADPAADAMARLDELSRQALQSREEVTAAQREVDAKLAEQSAAEER
MAB_0233|M.abscessus_ATCC_1997 LAITVATKAWAAVQALLNIALTANPIGLIIAAIAGLVAGIVVLYNRNETF
MMAR_0599|M.marinum_M KRRKPSVLLAA------------------------VVAPVAAFFAVAGDI
MUL_0559|M.ulcerans_Agy99 KRRKPSVLRAA------------------------VVAPAAAFFAVAGDI
Mb0328|M.bovis_AF2122/97 DRRKSSAVLAA------------------------VLAPAAVFFATGGDV
Rv0320|M.tuberculosis_H37Rv DRRKSSAVLAA------------------------VLAPAAVFFATGGDV
Mflv_1672|M.gilvum_PYR-GCK KRRNPSVYFVS------------------------ALAPAAVFLAVKGDT
Mvan_5081|M.vanbaalenii_PYR-1 KRRNPSVYVVS------------------------ALAPAAVFLAVKADT
MSMEG_0651|M.smegmatis_MC2_155 RRRKPSAVAAA------------------------VIAPAAVFFAVGGDV
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu HRADQEALAAANIRLQPHQ---------------EAVNKVAAMTYMGGST
MAB_0233|M.abscessus_ATCC_1997 RKIVQTTWAAIKTAISAVWNWLSTTVFPGLKLAFTAIGTAATWLWNNAIT
MMAR_0599|M.marinum_M APSS----------------------------------------------
MUL_0559|M.ulcerans_Agy99 GPSS----------------------------------------------
Mb0328|M.bovis_AF2122/97 STLA----------------------------------------------
Rv0320|M.tuberculosis_H37Rv STLA----------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK NPVSA---------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 SAIR----------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 NPFP----------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu DPISATLTATAPQQLIDRLAIQRMVTVQITE-------------------
MAB_0233|M.abscessus_ATCC_1997 PAWNGIKAVIGVAWEVVSDIFNNWVRVGQLVGQGAMWLWNNAIQPAWDGI
MMAR_0599|M.marinum_M ------------------------ARDRIDQMAV--------GDGACCVE
MUL_0559|M.ulcerans_Agy99 ------------------------ARDRIDQMAV--------GDEACCVE
Mb0328|M.bovis_AF2122/97 ------------------------ARADANPVLG--------DDAPCCVQ
Rv0320|M.tuberculosis_H37Rv ------------------------ARADANPVLG--------DDAPCCVQ
Mflv_1672|M.gilvum_PYR-GCK -----------------------SVAAEKEPVAAPAPPPVEEQATPCCVE
Mvan_5081|M.vanbaalenii_PYR-1 ------------------------VAVEEEPVAAPAPAPVEEQFVPCCVE
MSMEG_0651|M.smegmatis_MC2_155 ------------------------AEPDIKPAAAPEGT---GQPVPGMEL
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu ------------------RLASFRAERERAAGAARASERSAAEARAAAER
MAB_0233|M.abscessus_ATCC_1997 KNAISAAWDFVSPILDKFSAGWDALKSGISSASSAIKDAVTSAFSGLAAV
MMAR_0599|M.marinum_M IVA-----------------------------------------------
MUL_0559|M.ulcerans_Agy99 IVA-----------------------------------------------
Mb0328|M.bovis_AF2122/97 IVP-----------------------------------------------
Rv0320|M.tuberculosis_H37Rv IVP-----------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK IVA-----------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 VVA-----------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 IAA-----------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu AAAEKEQL------------------------------QAKWKDLLRQIA
MAB_0233|M.abscessus_ATCC_1997 IKAPLKLLGTFLASIPSEVFGFQIPGADKLNSWGKSLQGFAAGGLVRGPG
MMAR_0599|M.marinum_M ------ATPIELSSKISSGAVGMGFAAGQFAAASRLRVDRSARLLPVGVA
MUL_0559|M.ulcerans_Agy99 ------ATPIELSSKISSGAVGMGFAAGQFAAASRLRVDRSARLLPVGVA
Mb0328|M.bovis_AF2122/97 ------VAPLAFSSQISGGEIGTGLAASQFASASRWRIVS--RYLPVGVA
Rv0320|M.tuberculosis_H37Rv ------VAPLAFSSQISGGEIGTGLAASQFASASRWRIVS--RYLPVGVA
Mflv_1672|M.gilvum_PYR-GCK ------APATSPQPALLTDPGSAATSPASLVSASRWRVVDR--TLPAGVA
Mvan_5081|M.vanbaalenii_PYR-1 ------APASGPGPARLTDDGVADVEPASLVSASRWRVVSR--TLPAGVA
MSMEG_0651|M.smegmatis_MC2_155 ------GPAANPS---ATPAGVSAG--AEPATASRWRVLNIPKLLPAGRA
MAV_4829|M.avium_104 ---------------MASTRLSKGFGDGQPVAASRYHTRSR--FLPAGVA
TH_1403|M.thermoresistible__bu AAEAQYAALTPEQQQMVVNAAAPVPGAGSPLELAAARADLNLESLPVGVA
MAB_0233|M.abscessus_ATCC_1997 SGTSDSILAWLSNGEGVVTAKGMKHGAGIVAALNSGWVPSPAYLADMMRA
. *
MMAR_0599|M.marinum_M P--ERGLQVKTILAARSISAVFPEIHEIGGVRP-DALRWHPNGLALDVMI
MUL_0559|M.ulcerans_Agy99 P--ERGLQVKTILAARSISAVFPEIHEIGGVRP-DALRWHPNGLALDVMI
Mb0328|M.bovis_AF2122/97 P--EQGLQVKTVLTARSISAAFPEIREIGGVRP-DALRWHPNGLALDVMV
Rv0320|M.tuberculosis_H37Rv P--EQGLQVKTVLTARSISAAFPEIREIGGVRP-DALRWHPNGLALDVMV
Mflv_1672|M.gilvum_PYR-GCK P--ERGLQVRTILTSRSISAVFPEINNIGGVRS-DALRWHPNGLALDVMI
Mvan_5081|M.vanbaalenii_PYR-1 P--ENGLQVRTILTARSISAVFPEINNIGGVRQ-DALRWHPNGLALDVMI
MSMEG_0651|M.smegmatis_MC2_155 P--ESGLQVKTILAARSVSADFPEIQQIGGVRA-DSLRWHPNGLALDVMI
MAV_4829|M.avium_104 P--ERGLQVRTILTARSISAEFPQIHEIGGVRP-DPLPWHPLGMAIDVMI
TH_1403|M.thermoresistible__bu P--EAGLQPNTVLVARAISARFPQIAEIGGVRP-DSLPWHPSGLAIDVMI
MAB_0233|M.abscessus_ATCC_1997 PGYAEGLNPGADYLRSLVMRMWPQIKSIGGRRSEDGFGEHSTGNAIDIMI
* **: : : :*:* .*** * * : *. * *:*:*:
MMAR_0599|M.marinum_M PNPGSAEGIALGNEIVAYVLANAERFGMQDAIWRGVYYTPSGATRS----
MUL_0559|M.ulcerans_Agy99 PNPGSAEGIALGNEIVAYVLANTERFGMQDAIWRGVYYTPTGATRS----
Mb0328|M.bovis_AF2122/97 PNPGTAEGIALGNEIVAFVLKNATRFGMQDVIWRGAYYTPNGARTT----
Rv0320|M.tuberculosis_H37Rv PNPGTAEGIALGNEIVAFVLKNATRFGMQDVIWRGAYYTPNGARTT----
Mflv_1672|M.gilvum_PYR-GCK PNPTSQAGIALGNQIVAYVLANAKRFGIQDAIWRGTYYTPGGAQSG----
Mvan_5081|M.vanbaalenii_PYR-1 PNPTSQAGIALGNQIVAYVMANAERFGIQDAIWRGAYYTPGGAKAG----
MSMEG_0651|M.smegmatis_MC2_155 PNPSSPAGIALGNEIVAYALKNAERFALQDCIWRDTYYTPSGARKG----
MAV_4829|M.avium_104 PDPESAQGIALGNAIVAYVLQNADRFGIQDAIWRGVYYTPNGAQTS----
TH_1403|M.thermoresistible__bu PNHHSPEGIALGDEIMAFVLSNAARFGVQDVIWRGTYYTPSGPQGS----
MAB_0233|M.abscessus_ATCC_1997 PNYQTPQGMALGNAIAAFLANNASALDLNGFIWRRQSYGYGGSFTSGTPM
*: : *:***: * *: *: : ::. *** * *.
MMAR_0599|M.marinum_M ------RLGHYDHVHVTTTGGG---------------YPTGKEIYLR---
MUL_0559|M.ulcerans_Agy99 ------RLGHYDHVHVTTTGGG---------------YPTGKEIYLR---
Mb0328|M.bovis_AF2122/97 ------GAGHYDHIHITTVGGG---------------YPTGEELYIR---
Rv0320|M.tuberculosis_H37Rv ------GAGHYDHIHITTVGGG---------------YPTGEELYIR---
Mflv_1672|M.gilvum_PYR-GCK ------GYGHYDHVHVTTTGGG---------------YPDGDEVYYR---
Mvan_5081|M.vanbaalenii_PYR-1 ------GYGHYDHVHVTTTGGG---------------YPDGDEVYFR---
MSMEG_0651|M.smegmatis_MC2_155 ------SYGHYDHVHITTKGGG---------------YPSGGEVYLR---
MAV_4829|M.avium_104 ------RLGHYDHVHVTTIGGG---------------YPTGDEVYLR---
TH_1403|M.thermoresistible__bu ------GYGHFDHVHVTTLPGR----------------------------
MAB_0233|M.abscessus_ATCC_1997 DDRGDDNKNHMNHVHVILGSGRGSGAAAVGLPTSNISLPSGGSVAARAMG
.* :*:*: *
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 NTGSSGGSSKQAREADDRITDLSNRLDVTEQELADLESNPKAKETTKQRK
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 RDMVDKLKRDLQQAKDDRASLDLGGSSGGFGGGNNPYAKIMEGISEILPD
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 FGGLADIGIGGLKESLLPPGFSDPMQWGFMQAGSTLLKFFGGLRNVSDGK
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 PLLGEGGALMANITGAAMSGSGSGIVDAIKTIIPAPFGSMDAAQLQGAPG
MMAR_0599|M.marinum_M --------------------------------------------------
MUL_0559|M.ulcerans_Agy99 --------------------------------------------------
Mb0328|M.bovis_AF2122/97 --------------------------------------------------
Rv0320|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1672|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5081|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0651|M.smegmatis_MC2_155 --------------------------------------------------
MAV_4829|M.avium_104 --------------------------------------------------
TH_1403|M.thermoresistible__bu --------------------------------------------------
MAB_0233|M.abscessus_ATCC_1997 DINPVIAGAQIPGTGFGDMGSAFSQGSAAPNGNGSPVDQSINFNAPVGTG
MMAR_0599|M.marinum_M ---------------------------
MUL_0559|M.ulcerans_Agy99 ---------------------------
Mb0328|M.bovis_AF2122/97 ---------------------------
Rv0320|M.tuberculosis_H37Rv ---------------------------
Mflv_1672|M.gilvum_PYR-GCK ---------------------------
Mvan_5081|M.vanbaalenii_PYR-1 ---------------------------
MSMEG_0651|M.smegmatis_MC2_155 ---------------------------
MAV_4829|M.avium_104 ---------------------------
TH_1403|M.thermoresistible__bu ---------------------------
MAB_0233|M.abscessus_ATCC_1997 VDQAMQKAQSAQNQQYRRNNGTRTMPS