For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VEVERHSYDVVVIGAGGAGLRAVIEARERGLKVAVVCKSLFGKAHTVMAEGGCAAAMGNSNPKDNWKTHF GDTMRGGKFLNNWRMAELHAKEAPDRVWELETYGALFDRLKDGKISQRNFGGHTYPRLAHVGDRTGLELI RTMQQKIVSLQQEDFAEVGDYEARIKVFAECTITELLKDGPENGGRIAGAFGYWRESGRFILFEAPAVVM ATGGIGKSFKVTSNSWEYTGDGHALALRAGATLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKN SDDDRFMFGYIPPVFKGQYAETAEEADQWLKDNDSARRTPDLLPRDEVARAINSEVKAGRGTPHGGVYLD IASRLTPEEIKRRLPSMYHQFMELAGVDITKEPMEVGPTCHYVMGGIEVDADTGAATVPGLFAAGECSGG MHGSNRLGGNSLSDLLVFGRRAGLGAADYVRALSSRPTVSPESVDAAARLALAPFEGPTDGTAPENPYAL QMDLQFLMNDLVGIIRNADEISKALTLLRELWARYEHVQVEGHRQYNPGWNLAIDMRNMLTVSECVARAA LERTESRGGHTRDDHPGMDPNWRNVLLVCRAAQEATDGSHITITRQPQVPMRPDLLDLFQISELEKYYTA EELADHPGRRT*
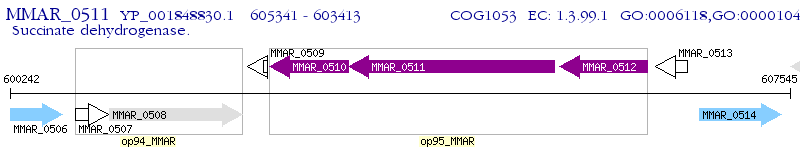
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0511 | sdhA | - | 100% (642) | succinate dehydrogenase (iron-sulfur subunit), SdhA_1 |
| M. marinum M | MMAR_1201 | sdhA | 2e-88 | 36.97% (633) | succinate dehydrogenase (flavoprotein subunit) SdhA |
| M. marinum M | MMAR_2392 | nadB | 5e-52 | 31.39% (599) | l-aspartate oxidase NadB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0254c | sdhA | 0.0 | 88.44% (649) | succinate dehydrogenase flavoprotein subunit |
| M. gilvum PYR-GCK | Mflv_0394 | sdhA | 0.0 | 76.24% (644) | succinate dehydrogenase flavoprotein subunit |
| M. tuberculosis H37Rv | Rv0248c | sdhA | 0.0 | 88.44% (649) | succinate dehydrogenase flavoprotein subunit |
| M. leprae Br4923 | MLBr_00697 | sdhA | 6e-89 | 36.20% (627) | succinate dehydrogenase flavoprotein subunit |
| M. abscessus ATCC 19977 | MAB_4422 | sdhA | 0.0 | 81.46% (642) | succinate dehydrogenase flavoprotein subunit |
| M. avium 104 | MAV_4909 | sdhA | 0.0 | 86.56% (640) | succinate dehydrogenase flavoprotein subunit |
| M. smegmatis MC2 155 | MSMEG_0418 | sdhA | 0.0 | 79.88% (641) | succinate dehydrogenase flavoprotein subunit |
| M. thermoresistible (build 8) | TH_4672 | - | 0.0 | 81.59% (641) | PUTATIVE succinate dehydrogenase subunit A |
| M. ulcerans Agy99 | MUL_1172 | sdhA | 0.0 | 99.69% (642) | succinate dehydrogenase flavoprotein subunit |
| M. vanbaalenii PYR-1 | Mvan_0284 | sdhA | 0.0 | 78.04% (633) | succinate dehydrogenase flavoprotein subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0511|M.marinum_M ----MVEVERHSYDVVVIGAGGAGLRAVIEARERGLKVAVVCKSLFGKAH
MUL_1172|M.ulcerans_Agy99 ----MVEVERHSYDVVVIGAGGAGLRAVIEARERGLKVAVVCKSLFGKAH
Mb0254c|M.bovis_AF2122/97 ----MVEVERHSYDVVVIGAGGAGLRAVIEARERGLKVAVVCKSLFGKAH
Rv0248c|M.tuberculosis_H37Rv ----MVEVERHSYDVVVIGAGGAGLRAVIEARERGLKVAVVCKSLFGKAH
MAV_4909|M.avium_104 ----MVEVERHAYDVVVIGAGGAGLRAVIEARERGLRVAVVCKSLFGKAH
TH_4672|M.thermoresistible__bu ----MTEVERHQYDVVVIGAGGSGLRAVIEARERGLRVAVVTKSLFGKAH
Mflv_0394|M.gilvum_PYR-GCK MASEEAQLERHEYDVVVIGAGGAGLRAVIEARERGLRVAVVTKSLFGKAH
Mvan_0284|M.vanbaalenii_PYR-1 --------------MVVIGAGGAGLRAVIEARERGLRVAVVTKSLFGKAH
MSMEG_0418|M.smegmatis_MC2_155 ----MSELERHSYDVVVIGAGGAGLRAVIEARERGLRVAVVTKSLFGKAH
MAB_4422|M.abscessus_ATCC_1997 ----MAELERHQYDVVVIGAGGAGLRAVIEAREQGFKVAVVCKSLFGKAH
MLBr_00697|M.leprae_Br4923 ------MIQQHRYDVVIVGAGGAGMRAAVEAGPR-VRTAVLTKLYPTRSH
:*::****:*:**.:** : .:.**: * ::*
MMAR_0511|M.marinum_M TVMAEGGCAAAMGNSNPKDNWKTHFGDTMRGGKFLNNWRMAELHAKEAPD
MUL_1172|M.ulcerans_Agy99 TVMAEGGCAAAMGNNNPKDNWKTHFGDTMRGGKFLNNWRMAELHAKEAPD
Mb0254c|M.bovis_AF2122/97 TVMAEGGCAAAMGNANPKDNWKTHFGDTMRGGKFLNNWRMAELHAKEAPD
Rv0248c|M.tuberculosis_H37Rv TVMAEGGCAAAMGNANPKDNWKTHFGDTMRGGKFLNNWRMAELHAKEAPD
MAV_4909|M.avium_104 TVMAEGGCAASMGNTNPKDNWKTHFGDTMRGGKFLNNWRMAELHAKEAPD
TH_4672|M.thermoresistible__bu TVMAEGGCAASMGNTNPKDNWQTHFCDTMRGGKFLNNWRMAELHAKESPD
Mflv_0394|M.gilvum_PYR-GCK TVMAEGGCAAAMRNVNTKDSWQVHFGDTMRGGKFLNNWRMAELHAQEAPD
Mvan_0284|M.vanbaalenii_PYR-1 TVMAEGGCAAAMRNVNTKDSWQVHFGDTMRGGKFLNNWRMAELHAQEAPD
MSMEG_0418|M.smegmatis_MC2_155 TVMAEGGCAAAMRNVNTKDSWQVHFGDTMRGGKFLNNWRMAELHAQEAPD
MAB_4422|M.abscessus_ATCC_1997 TVMAEGGCAASMGNANDKDSWQIHFGDTMRGGKFLNNWRMAELHAKEAPD
MLBr_00697|M.leprae_Br4923 TGAAQGGMCAALANVED-DNWEWHAFDTVKGGDYLADQDAVEIMCKEAID
* *:** .*:: * : *.*: * **::**.:* : .*: .:*: *
MMAR_0511|M.marinum_M RVWELETYGALFDRLKDGKISQRNFGGHT-------YPRLAHVGDRTGLE
MUL_1172|M.ulcerans_Agy99 RVWELETYGALFDRLKDGKISQRNFGGHT-------YPRLAHVGDRTGLE
Mb0254c|M.bovis_AF2122/97 RVWELETYGALFDRTDDGRISQRNFGGHT-------YPRLAHVGDRTGLE
Rv0248c|M.tuberculosis_H37Rv RVWELETYGALFDRTDDGRISQRNFGGHT-------YPRLAHVGDRTGLE
MAV_4909|M.avium_104 RVWELETYGALFDRLKDGKISQRNFGGHT-------YPRLAHVGDRTGLE
TH_4672|M.thermoresistible__bu RVWELETYGALFDRLEDGRISQRNFGGHT-------YPRLAHVGDRTGLE
Mflv_0394|M.gilvum_PYR-GCK RVWELETYGALFDRTKDGRISQRNFGGHT-------YPRLAHVGDRTGLE
Mvan_0284|M.vanbaalenii_PYR-1 RVWELETYGALFDRTKDGRISQRNFGGHT-------YPRLAHVGDRTGLE
MSMEG_0418|M.smegmatis_MC2_155 RVWELETYGALFDRTKDGKISQRNFGGHT-------YPRLAHVGDRTGLE
MAB_4422|M.abscessus_ATCC_1997 RVWELETYGALFDRTKDGRISQRNFGGHT-------YPRLAHVGDRTGLE
MLBr_00697|M.leprae_Br4923 AVLDLEKMGMPFNRTPEGRIDQRRFGGHTRDHGKAPVRRACYAADRTGHM
* :**. * *:* :*:*.**.***** * .:..****
MMAR_0511|M.marinum_M LIRTMQQKIVSLQQEDFAEVGDYEARIKVFAECTITELLKDGPENGGRIA
MUL_1172|M.ulcerans_Agy99 LIRTMQQKIVSLQQEDFAEVGDYEARIKVFAECTITELLKDGPENGGRIA
Mb0254c|M.bovis_AF2122/97 LIRTLQQKVVSLQQEDHAELGDYEARIKVFAECTITELLKDQ----GAIA
Rv0248c|M.tuberculosis_H37Rv LIRTLQQKVVSLQQEDHAELGDYEARIKVFAECTITELLKDQ----GAIA
MAV_4909|M.avium_104 LIRTMQQKIVSLQQEDYAELGDYEARIKVFAECTITELLKDG----DAIA
TH_4672|M.thermoresistible__bu LIRTMQQKIVSLQQEDKAEFGDYEARIRVFHECSITELIKDG----DRIA
Mflv_0394|M.gilvum_PYR-GCK IIRTLQQKIVSLQQEDKRELGDFDARIRVFHECSITEIIKEN----GRVA
Mvan_0284|M.vanbaalenii_PYR-1 IIRTLQQKIVSLQQEDKKELGDFEARIRVFHETSITELIKDG----DRIA
MSMEG_0418|M.smegmatis_MC2_155 IIRTLQQKIVSLQQEDKRELGDYEARIRVFHETSITELILDD----GKIA
MAB_4422|M.abscessus_ATCC_1997 IIRTMQQKIVSLQQEDFAETGDYEARIKIFAECTITDLLLDSTDGVSRIS
MLBr_00697|M.leprae_Br4923 ILQTLYQNCVKHDVEFFNEFYALDLVLTQTPSGPVT-------------T
:::*: *: *. : * * : : . .:* :
MMAR_0511|M.marinum_M GAFGYWRESGRFILFEAPAVVMATGGIGKSFKVTSNSWEYTGDGHALALR
MUL_1172|M.ulcerans_Agy99 GAFGYWRESGRFILFEAPAVVMATGGIGKSFKVTSNSWEYTGDGHALALR
Mb0254c|M.bovis_AF2122/97 GAFGYWRESGRFIVFEAPAVVLATGGIGKSFKVTSNSWEYTGDGHALALR
Rv0248c|M.tuberculosis_H37Rv GAFGYWRESGRFIVFEAPAVVLATGGIGKSFKVTSNSWEYTGDGHALALR
MAV_4909|M.avium_104 GAFGYWRESGQFVLFEAPAVVLATGGIGKSFKVTSNSWEYTGDGHALALR
TH_4672|M.thermoresistible__bu GAFGYWRETGRFILFEAPAVVLATGGIGKTYKVTSNSWEYTGDGHALALR
Mflv_0394|M.gilvum_PYR-GCK GAFGYYRETGKFVLFEAPAIVLATGGIGKSFKVSSNSWEYTGDGHALALR
Mvan_0284|M.vanbaalenii_PYR-1 GAFGYYRETGKFVLFEAPAIVLATGGIGKSFKVSSNSWEYTGDGHALALR
MSMEG_0418|M.smegmatis_MC2_155 GAFGYYRETGNFVLFEAPAVVLATGGIGKSFKVSSNSWEYTGDGHALALR
MAB_4422|M.abscessus_ATCC_1997 GAFGYWRESGRFILFEAPAVVLATGGIGKSFKVSSNSWEYTGDGHALALR
MLBr_00697|M.leprae_Br4923 GVVAYELATGDIHVFHTKAVVIATGGSGRMYKTTSNAHTLTGDGIGIVFR
*...* :* : :*.: *:*:**** *: :*.:**: **** .:.:*
MMAR_0511|M.marinum_M AGATLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSDDDRFMF
MUL_1172|M.ulcerans_Agy99 AGATLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSDDDRFMF
Mb0254c|M.bovis_AF2122/97 AGATLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSENSRFMF
Rv0248c|M.tuberculosis_H37Rv AGATLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSENSRFMF
MAV_4909|M.avium_104 AGASLINMEFVQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSEGKRFMF
TH_4672|M.thermoresistible__bu AGSTLINMEFIQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSEGKRFMF
Mflv_0394|M.gilvum_PYR-GCK AGSGLINMEFIQFHPTGMVWPLSVKGILVTEGVRGDGGVLKNSEGKRFMF
Mvan_0284|M.vanbaalenii_PYR-1 AGSGLINMEFIQFHPTGMVWPLSVKGILVTEGVRGDGGVLKNSDGKRFMF
MSMEG_0418|M.smegmatis_MC2_155 AGSALINMEFIQFHPTGMVWPLSVKGILVTEGVRGDGGVLKNSEGKRFMF
MAB_4422|M.abscessus_ATCC_1997 AGGTLINMEFIQFHPTGMVWPPSVKGILVTEGVRGDGGVLKNSEGKRFMF
MLBr_00697|M.leprae_Br4923 KGLPLEDMEFHQFHPTGLAG----LGILISEAVRGEGGRLLNGENERFME
* * :*** ******:. ***::*.***:** * *.:..***
MMAR_0511|M.marinum_M GYIPPVFKGQYAETAEEADQWLKDNDSARRTPDLLPRDEVARAINSEVKA
MUL_1172|M.ulcerans_Agy99 GYIPPVFKSQYAETAEEADQWLKDNDSARRTPDLLPRDEVARAINSEVKA
Mb0254c|M.bovis_AF2122/97 DYIPPVFKGQYAETEEEADQWLKDNDSARRTPDLLPRDEVARAINSEVKA
Rv0248c|M.tuberculosis_H37Rv DYIPPVFKGQYAETEEEADQWLKDNDSARRTPDLLPRDEVARAINSEVKA
MAV_4909|M.avium_104 DYIPPVFKGQYAETEQEADQWLKDNDSARRTPDLLPRDEVARAINSEVKA
TH_4672|M.thermoresistible__bu DYIPAVFKGQYAETEEEADQWLKDNDSARRTPDLLPRDEVARAINAEVKA
Mflv_0394|M.gilvum_PYR-GCK DYIPDVFKGQYAESEEEADQWLKDNDSARRTPDLLPRDEVARAINEEVKN
Mvan_0284|M.vanbaalenii_PYR-1 DYIPEVFKGQYAESEEEADQWLKDNDSARRTPDLLPRDEVARAINEEVKA
MSMEG_0418|M.smegmatis_MC2_155 DYIPSVFKGQYAETEEEADQWLKDNDSARRTPDLLPRDEVARAINAEVKA
MAB_4422|M.abscessus_ATCC_1997 DYIPSVFKGQYAETEEEADQWLKDNDSARRTPDLLPRDEVARAINAEVKA
MLBr_00697|M.leprae_Br4923 HYAPTIV-------------------------DLAPRDIVARSMVLEVLE
* * :. ** *** ***:: **
MMAR_0511|M.marinum_M GRGT-PHGG-VYLDIASRLTPEEIKRRLPSMYHQFMELAGVDITKEPMEV
MUL_1172|M.ulcerans_Agy99 GRGT-PHGG-VYLDIASRLTPEEIKRRLPSMYHQFMELAGVDITKEPMEV
Mb0254c|M.bovis_AF2122/97 GRGT-PHGG-VYLDIASRLTPAEIKRRLPSMYHQFKELAEVDITTQAMEV
Rv0248c|M.tuberculosis_H37Rv GRGT-PHGG-VYLDIASRLTPAEIKRRLPSMYHQFKELAEVDITTQAMEV
MAV_4909|M.avium_104 GRGS-PHGG-VFLDIASRLTPAEINRRLPSMYHQFKELAGVDITKEPMEV
TH_4672|M.thermoresistible__bu GRGS-PHGG-VYLDIASRLPEGEILKRLPSMYHQFKELAEVDITKEPMEV
Mflv_0394|M.gilvum_PYR-GCK GRGT-PHGG-VYLDIASRMPAEEIKRRLPSMYHQFIELAEVDITKDEMEV
Mvan_0284|M.vanbaalenii_PYR-1 GRGT-PHGG-VYLDIASRMPAEEIKRRLPSMYHQFIELAEVDITKDEMEV
MSMEG_0418|M.smegmatis_MC2_155 GRGS-PHGG-VYLDIASRMPAEEIKRRLPSMYHQFIELAEVDITKDAMEV
MAB_4422|M.abscessus_ATCC_1997 GRGT-PHGG-VYLDIASRIPTEEIKKRLPSMYHQFKELAEVDITKDDMEV
MLBr_00697|M.leprae_Br4923 GRGAGPHKDYVYIDVR-HLGEEVLESKLPDITEFSRTYLGVDPVHELVPV
***: ** . *::*: :: : :**.: . ** . : : *
MMAR_0511|M.marinum_M GPTCHYVMGGIEVDAD-----TGAATVPGLFAAGECSGG-MHGSNRLGGN
MUL_1172|M.ulcerans_Agy99 GPTCHYVMGGIEVDAD-----TGAATVPGLFAAGECSGG-MHGSNRLGGN
Mb0254c|M.bovis_AF2122/97 GPTCHYVMGGVEVDAD-----TGAATVPGLFAAGECAGG-MHGSNRLGGN
Rv0248c|M.tuberculosis_H37Rv GPTCHYVMGGVEVDAD-----TGAATVPGLFAAGECAGG-MHGSNRLGGN
MAV_4909|M.avium_104 GPTCHYVMGGVEVDAD-----TGAATVPGLFAAGECSGG-MHGSNRLGGN
TH_4672|M.thermoresistible__bu GPTCHYIMGGIEVDPD-----PAAARTPGLFAAGECAGG-MHGSNRLGGN
Mflv_0394|M.gilvum_PYR-GCK GPTCHYVMGGIEVDPD-----TGEAATPGLFAAGECSGG-MHGSNRLGGN
Mvan_0284|M.vanbaalenii_PYR-1 GPTCHYVMGGIEVDPD-----SGGAATLGLFAAGECSGG-MHGSNRLGGN
MSMEG_0418|M.smegmatis_MC2_155 GPTCHYVMGGIEVDPD-----TAAGATPGLFAAGECSGG-MHGSNRLGGN
MAB_4422|M.abscessus_ATCC_1997 GPTCHYVMGGIEVDPD-----SGAATVPGLFAAGECSGG-MHGSNRLGGN
MLBr_00697|M.leprae_Br4923 YPTCHYVMGGIPTTVTGQVLRDNTSTVPGLYAAGECACVSVHGANRLGTN
*****:***: . . . **:*****: :**:**** *
MMAR_0511|M.marinum_M SLSDLLVFGRRAGLGAADYVRA--LSSRPTVSPESVDAAARLALAPFEGP
MUL_1172|M.ulcerans_Agy99 SLSDLLVFGRRAGLGAADYVRA--LSSRPTVSPESVDAAARLALAPFEGP
Mb0254c|M.bovis_AF2122/97 SLSDLLVFGRRAGLGAADYVRA--LSSRPAVSAEAIDAAAQQALSPFEGP
Rv0248c|M.tuberculosis_H37Rv SLSDLLVFGRRAGLGAADYVRA--LSSRPAVSAEAIDAAAQQALSPFEGP
MAV_4909|M.avium_104 SLSDLLVFGRRAGLGAADYVRA--LSSRPTISPDAVEAAAKRALAPFEAP
TH_4672|M.thermoresistible__bu SLSDLLVFGRRAGLGAADYVNA--LKDRPQVSQEAVDHAIRLALAPFEGP
Mflv_0394|M.gilvum_PYR-GCK SLSDLLVFGRRAGLGASDYVRS--LDNRPAVTDAALQRAEQLALLPFEPK
Mvan_0284|M.vanbaalenii_PYR-1 SLSDLLVFGRRAGLGASDYVRS--LPDRPKVSEVALERAEKLALEPFEAK
MSMEG_0418|M.smegmatis_MC2_155 SLSDLLVFGRRAGLGAADYVRA--LPDRPKVSEAAVEDATRLVLAPFEPK
MAB_4422|M.abscessus_ATCC_1997 SLSDLLVFGRRAGLGAADYVKS--LTSRPQITEADVDKAAALALAPFGDP
MLBr_00697|M.leprae_Br4923 SLLDINVFGRRSGIAAASYASANDFVDMPPNPAEMVISWVANILSEHGN-
** *: *****:*:.*:.*. : : . * . : * .
MMAR_0511|M.marinum_M TDGTAPENPYALQMDLQFLMNDLVGIIRNADEISKALTLLRELWARYEHV
MUL_1172|M.ulcerans_Agy99 TDGTAPENPYALQMDLQFLMNDLVGIIRNADEISKALTLLRELWARYEHV
Mb0254c|M.bovis_AF2122/97 KDGSAPENPYALHMDLQYVMNDLVGIIRNADEISRALTLLAELWSRYHNV
Rv0248c|M.tuberculosis_H37Rv KDGSAPENPYALHMDLQYVMNDLVGIIRNADEISRALTLLAELWSRYHNV
MAV_4909|M.avium_104 AGGTA-ENPYTLQLELQQSMNDLVGIIRNADEISEALARLGKLRERFKNL
TH_4672|M.thermoresistible__bu SNGATPENPYTLQLDLQDTMNELVGIIRTEAEMTQALNKLGELRERFRNL
Mflv_0394|M.gilvum_PYR-GCK ADA---ENPYTLHAELQESMNDLAGIIRKENELHEVLAKIDELKKRYQNV
Mvan_0284|M.vanbaalenii_PYR-1 DNA---ENPYTLHAELQESMNDLAGIIRKQNELEEVLLKIDELKRRYRNV
MSMEG_0418|M.smegmatis_MC2_155 AEP---ENPYTLHAELQQSMNDLVGIIRKEAEIQEALDRLQELKRRYANV
MAB_4422|M.abscessus_ATCC_1997 AAAGA-ENPYTLHTDLQQAMNDLVGIIRKEEEVEQALVKLNELKERFKHV
MLBr_00697|M.leprae_Br4923 ------ERVADIRGALQQTMDNNAAVFRTEETLKQALTDIHSLKERYSRI
*. :: ** *:: ..::*. : ..* : .* *: .:
MMAR_0511|M.marinum_M QVEG-HRQYNPGWNLAIDMRNMLTVSECVARAALERTESRGGHTRDDHPG
MUL_1172|M.ulcerans_Agy99 QVEG-HRQYNPGWNLAIDMRNMLTVSECVARAALERTESRGGHTRDDHPG
Mb0254c|M.bovis_AF2122/97 LVEG-HRQYNPGWNLSIDLRNMLLVSECVARAALQRTESRGGHTRDDHPG
Rv0248c|M.tuberculosis_H37Rv LVEG-HRQYNPGWNLSIDLRNMLLVSECVARAALQRTESRGGHTRDDHPG
MAV_4909|M.avium_104 HVDG-QRRYNPGWNLALDLRNMLLVSECVAKAALERTESRGGHTRDDHPS
TH_4672|M.thermoresistible__bu KVDG-GRHYNPGWHLAIDLRNMLLVSECVAKAALDRTESRGGHTRDDYPA
Mflv_0394|M.gilvum_PYR-GCK VVEG-GRIFNPGWHLAIDMRNMLLVSECVAKAALQRTESRGGHTRDDYPK
Mvan_0284|M.vanbaalenii_PYR-1 VVEG-GRIFNPGWHLAIDMRNMLLVSECVAKAALKRTESRGGHTRDDFPQ
MSMEG_0418|M.smegmatis_MC2_155 TVEG-GRVFNPGWHLAIDMRNMLLVSECVAKAALQRTESRGGHTRDDYPE
MAB_4422|M.abscessus_ATCC_1997 AVEG-HRQFNPGWHLAIDMRNMLLVSECVAKAALLRTESRGGHTRDDHPN
MLBr_00697|M.leprae_Br4923 TVHDKGKSFNSDLLEAIELGFLLELAEVTVVGALNRKESRGGHAREDYPH
*.. : :*.. :::: :* ::* .. .** *.******:*:*.*
MMAR_0511|M.marinum_M MDPNWRNVLLVCRAAQEATDG--------SHITITRQPQVPMRPDLLDLF
MUL_1172|M.ulcerans_Agy99 MDPNWRNVLLVCRAAQEATDG--------SHITITRQPQVPMRPDLLDLF
Mb0254c|M.bovis_AF2122/97 MDPNWRRILLVCRATETMGTGGSGSGDSNCHINVTQQLQTPMRPDLLELF
Rv0248c|M.tuberculosis_H37Rv MDPNWRRILLVCRATETMGTGGSGSGDSNCHINVTQQLQTPMRPDLLELF
MAV_4909|M.avium_104 MDSAWRKVLLVCEAAGGDEVIP--------DISIRKKDQTPMRPDLLELF
TH_4672|M.thermoresistible__bu MDAKWRSTLLVCRATGDDDVLP--------SIEVTPEQQPPMRPDLLALF
Mflv_0394|M.gilvum_PYR-GCK MDSHWRNKLLVCKTVGGGDEPG---DPIIPEVSVEVEPQPPMRPDLLATF
Mvan_0284|M.vanbaalenii_PYR-1 MDSNWRNKLLVCKTVGGGD------DPVIPEISVEVEPQPPMRPDLLATF
MSMEG_0418|M.smegmatis_MC2_155 MDANWRNTLLVCR-VSGGD-------PVVPDVTVTPEQQVPMRPDLLGCF
MAB_4422|M.abscessus_ATCC_1997 MDATWRKTLLVCS-TTGGD-------PAVPDVLVTPEPQLPLRPDLLALF
MLBr_00697|M.leprae_Br4923 RDDTN-----------------------YMRHTMAYKQGTDLLSDIALDF
* : : : .*: *
MMAR_0511|M.marinum_M QISELEKYYTAEELADHPGR--RT-------
MUL_1172|M.ulcerans_Agy99 QISELEKYYTAEELADHPGR--RT-------
Mb0254c|M.bovis_AF2122/97 EISELEKYYTDEELAEHPGR--RG-------
Rv0248c|M.tuberculosis_H37Rv EISELEKYYTDEELAEHPGR--RG-------
MAV_4909|M.avium_104 DIAELEKYYTDEELAGHPGRGPRTDPGTESK
TH_4672|M.thermoresistible__bu EIDELEKYYTDEELAEHPGR--RV-------
Mflv_0394|M.gilvum_PYR-GCK ELSELEKYYTPEELAEHPEQ--KG-------
Mvan_0284|M.vanbaalenii_PYR-1 ELSELEKYYTAEELAEHPQR--KG-------
MSMEG_0418|M.smegmatis_MC2_155 ELSELEKYYTPEELAEHPER--KG-------
MAB_4422|M.abscessus_ATCC_1997 EIGELEKYFTPEELAEHPGR--KS-------
MLBr_00697|M.leprae_Br4923 KPVVQTRYEPQERKY----------------
. :* . *.