For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AADKPNFWQYIAYSYGKCLPDSMRSWVANDLAGKGAIRRHMIRWALPPILILAPLWLLPASLYVHSEMTV PLYIWSLLISLALNKVWRRHRLAQHGLDPNLVDVLKQKKQARMHEDYARRYGPRPEEAHWQSNSSPF*
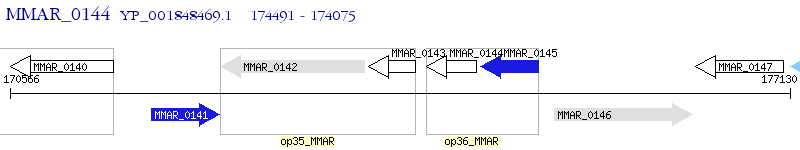
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0144 | - | - | 100% (138) | hypothetical protein MMAR_0144 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1909 | - | 7e-61 | 72.59% (135) | hypothetical protein Mflv_1909 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1030 | - | 5e-56 | 70.80% (137) | hypothetical protein MAB_1030 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0537 | - | 1e-61 | 77.78% (135) | hypothetical protein MSMEG_0537 |
| M. thermoresistible (build 8) | TH_0332 | - | 3e-42 | 56.82% (132) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4953 | - | 4e-80 | 98.55% (138) | hypothetical protein MUL_4953 |
| M. vanbaalenii PYR-1 | Mvan_4824 | - | 4e-58 | 69.63% (135) | hypothetical protein Mvan_4824 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0144|M.marinum_M ---------MAADKPNFWQYIAYSYGKCLPDSMRSWVANDLAGKGAIRRH
MUL_4953|M.ulcerans_Agy99 ---------MAADKPNFWQYIAYSYGKCLPDSMRSWVANDLAGKGAIRRH
Mflv_1909|M.gilvum_PYR-GCK ---MAHPEPTGTEKPSFWQYLAYSYGKRLPKSMDRWVAEDLAGEGAVRRH
Mvan_4824|M.vanbaalenii_PYR-1 ---MPLDEPQGTDKPTMWQYLTYCYGRRLPASMNRWVAEDLAGDGAVRRH
MSMEG_0537|M.smegmatis_MC2_155 -----MAAERKKDRPNLLQYIAYSYGRRLPDSMREWVAHDLADHGAVRRH
MAB_1030|M.abscessus_ATCC_1997 ---------MAADRPNLIQYIAYCYGKRLPDSMRDWVAHDLAGKGAVRRH
TH_0332|M.thermoresistible__bu VSNGPERATAGRTRPNAWEYITYCYGRRLPDSMRDWVRQDLAGKGAVRRM
:*. :*::*.**: ** ** ** .***..**:**
MMAR_0144|M.marinum_M MIRWALPPILILAPLWLLPASLYVHSEMTVPLYIWSLLISLALNKVWRRH
MUL_4953|M.ulcerans_Agy99 MIRWALPAILILAPLWLLPASLYVHSEMTVPLYIWSLLISLALNKVWRRH
Mflv_1909|M.gilvum_PYR-GCK MIRWAIPPLIVLAPLWLLPAPIYMRLEMTAPIYIWALLMALALNKVWRRH
Mvan_4824|M.vanbaalenii_PYR-1 MIRYAIPPLLILAPLWLIPAPIYMRLEMTAPIYIWALLMALALNKVWRRH
MSMEG_0537|M.smegmatis_MC2_155 MIRMAIPPALVLAPFWLLPASLYVHIEMTVPIYIWSLLMALALNKVWRRH
MAB_1030|M.abscessus_ATCC_1997 ILRCAIPPLFILAPFWLLPASLYVHMEMTVPIYVWVLIMAHALNKVWRRH
TH_0332|M.thermoresistible__bu MTRMFIPAFLVLAPFWLIDTTLYVKFSMTMPILVWAVLFSHALNKVWRRH
: * :*. ::***:**: :.:*:: .** *: :* :::: *********
MMAR_0144|M.marinum_M RLAQHGLDPNLVDVLKQKKQARMHEDYARRYGPRPEEAHWQSNSSPF
MUL_4953|M.ulcerans_Agy99 RLAQHGLDPDLVDVLKQKKQARMHEDYARRYGPRPEEAHWQSNSSPF
Mflv_1909|M.gilvum_PYR-GCK RLARHGLDPNLVDVIKRKKDAKMHDYYIRKYGPRPEDSKTQSNSSPF
Mvan_4824|M.vanbaalenii_PYR-1 RLAKHGMDPNLVDVIKRKRDAQMHEDYVRRFGPRPDDAQAHSNSSPF
MSMEG_0537|M.smegmatis_MC2_155 RLAQHGLDPNLVDEIRYKKQAHIHEDYIRRFGPRPESAKFQSNSSPF
MAB_1030|M.abscessus_ATCC_1997 RLVQHDLDPGLVDVLKRQKDARIHEDYARRFGPRSGDG--YSSSGPI
TH_0332|M.thermoresistible__bu MLRMHNLDPNLVDEIRRQKTAHIDRAYIAKYGPRPADA--QSNSSAV
* *.:**.*** :: :: *::. * ::***. .. *.*...