For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRSLAAVVGSSHVIADPDVLIAHSIDHTGRYRGQARALVRPGTAEQVTEVLRVCRDVGAHVTVQGGRTSL VAGTVPEHNDVLLSTERLRSISEVDTVERRLRVGAGATLTAVQNAATAAGLVFGVDLCARATATVGGMAS TNAGGLRTVRYGNMGEQVIGLEVALPDGSLLCRHSLARFDNTGYNLPALFVGAEGTLGVITALDLRLHPA PAHRVTAVCGFSDLGALVDASRAFRDVYGIAALELIDGRASSLIREHLGVSSPVDGDWLLLVELAADHDQ TNRLAELSMAVQMCGVPAVGVDAGAQQRLWRVRESLAEVLGVYGPPLKFDVSLPLSAISGFAKEAATLID AQVSDVQEALPVLFGHVGEGNLHLNVLRCPPDREKTLTAAMMELIAQAGGNVSSEHGVGSNKRAYLRMSR DPADIAAMRTIKAALDPTGYLNAAVLFD*
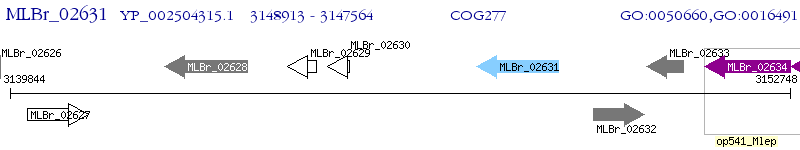
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02631 | - | - | 100% (449) | putative oxidoreductase |
| M. leprae Br4923 | MLBr_01103 | - | 5e-37 | 28.85% (454) | putative oxidoreductase subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0166 | - | 0.0 | 78.84% (449) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_2230 | - | 1e-39 | 30.33% (455) | FAD linked oxidase domain-containing protein |
| M. tuberculosis H37Rv | Rv0161 | - | 0.0 | 78.84% (449) | oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1398c | - | 1e-151 | 62.50% (448) | putative oxidoreductase |
| M. marinum M | MMAR_0388 | - | 0.0 | 78.84% (449) | oxidoreductase |
| M. avium 104 | MAV_5135 | - | 0.0 | 76.61% (449) | FAD linked oxidase |
| M. smegmatis MC2 155 | MSMEG_5045 | - | 0.0 | 73.42% (444) | D-2-hydroxyglutarate dehydrogenase |
| M. thermoresistible (build 8) | TH_0754 | - | 1e-177 | 70.76% (448) | D-2-hydroxyglutarate dehydrogenase |
| M. ulcerans Agy99 | MUL_1038 | - | 0.0 | 78.62% (449) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4473 | - | 1e-179 | 71.21% (448) | FAD linked oxidase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0166|M.bovis_AF2122/97 ----MLTSLVSAVGSHHVTTDPDVLAGRSVDHTGR-YRGRASALVRPGSA
Rv0161|M.tuberculosis_H37Rv ----MLTSLVSAVGSHHVTTDPDVLAGRSVDHTGR-YRGRASALVRPGSA
MMAR_0388|M.marinum_M ----MLRSLANVVGSSHVLTDPDVLAGRSVDHTGR-YRGRASALVRPASA
MUL_1038|M.ulcerans_Agy99 ----MLRSLANVVGSSHVLTDPDVLAGRSVDHTGR-YRGRASALVRPASA
MAV_5135|M.avium_104 ----MLRALADTVGSTHVVTDPDVLAARSVDHTGR-YRGRASALVRPGSA
MLBr_02631|M.leprae_Br4923 ----MLRSLAAVVGSSHVIADPDVLIAHSIDHTGR-YRGQARALVRPGTA
MSMEG_5045|M.smegmatis_MC2_155 MAEALTERLAHIVGAGHVSTDPDVLAARSVDHTGR-YRGRACALVRPGTG
Mvan_4473|M.vanbaalenii_PYR-1 MTEALIDRLAAIVGATQVSIDADVLEGRSVDHTGR-YRGHASALVRPGSA
TH_0754|M.thermoresistible__bu MTATLAGRLAQIVGPAHVSTDPDVLAGRSVDHTGR-YRGRATALVRPGDT
MAB_1398c|M.abscessus_ATCC_199 -MSTLARALVQIVGERYVSTDADVLAGRTIDHTGR-YLGTASILIRPGTA
Mflv_2230|M.gilvum_PYR-GCK --MQPLASLIADLPDGTVVTDPDILESYRQDRAADPAAGTPLAVVRPRRT
* : * *.*:* . *::. * . ::**
Mb0166|M.bovis_AF2122/97 EEVAEVLRVCRDAGAYVTVQGGRTSLVAGTVPEHDDVLLSTERLCVVSDV
Rv0161|M.tuberculosis_H37Rv EEVAEVLRVCRDAGAYVTVQGGRTSLVAGTVPEHDDVLLSTERLCVVSDV
MMAR_0388|M.marinum_M DQVAEVLRVCRDAGAHVTVQGGRTSLVAGTVPEHDDVLLSTERICDVADV
MUL_1038|M.ulcerans_Agy99 DQVAEVLRVCRDAGAHVTVQGGRTSLVAGTVPEHDDVLLSTERICDVADV
MAV_5135|M.avium_104 EQLAEVLRVCRDAGAHVTVQGGRTSLVAGTVPEHDDVLLSTERLHRIADV
MLBr_02631|M.leprae_Br4923 EQVTEVLRVCRDVGAHVTVQGGRTSLVAGTVPEHNDVLLSTERLRSISEV
MSMEG_5045|M.smegmatis_MC2_155 DQVAAVLRACRDAGAYVTVQGGRTSLVAGTVPEHDDVLLSTERLTTIGDV
Mvan_4473|M.vanbaalenii_PYR-1 DEVAAVLRVCRDAGVNVTVQGGRTSLVAGTVPEHDDVLLSTERLRDVGEV
TH_0754|M.thermoresistible__bu AEVAAVLRTCRDAGSHVTVQGGRTSLVAGTVPEHDDVLLSTERLTDIGPV
MAB_1398c|M.abscessus_ATCC_199 EEVAEILKVCKERSQAVTTQGGRTSMVAGTVSEHDDVLLSTERLTEIGPI
Mflv_2230|M.gilvum_PYR-GCK EEVQTVLRWAGTHRIAVVPRGMGTGLSGGATALDGGIVLSTEKMRDIT-V
:: :*: . *. :* *.: .*:.. ...::****:: : :
Mb0166|M.bovis_AF2122/97 DTVERRIEIGAGVTLAAVQHAASTAGLVFGVDLSARDTATVGGMASTNAG
Rv0161|M.tuberculosis_H37Rv DTVERRIEIGAGVTLAAVQHAASTAGLVFGVDLSARDTATVGGMASTNAG
MMAR_0388|M.marinum_M DTLERRVAVGAGATLAAVQRAATAAGLVFGVDLSARESATVGGMASTNAG
MUL_1038|M.ulcerans_Agy99 DTLERRVAVGAGATLAAVQRAATAAGLVFGVDLSARESATVGGMASTNAG
MAV_5135|M.avium_104 DTVERRIEVGAGATLAAVQQAASAANLVFGVDLAARDSATVGGMASTNAG
MLBr_02631|M.leprae_Br4923 DTVERRLRVGAGATLTAVQNAATAAGLVFGVDLCARATATVGGMASTNAG
MSMEG_5045|M.smegmatis_MC2_155 DTVERRISVGAGVTLAAVQKTAAAAGLVFGVDLAARDSATVGGMASTNAG
Mvan_4473|M.vanbaalenii_PYR-1 DVVERRIRVGAGATLAEVQRAAAAAGLVFGVDLAARDSATVGGMASTNAG
TH_0754|M.thermoresistible__bu DTVERRVWAGAGVTLAALQHTAAAVGLTFGVDLAARDTATVGGMAATNAG
MAB_1398c|M.abscessus_ATCC_199 DTVERRVRAGSGVTLAALQQAAAKENLQFGVDIGSRDSATLGGMASTNAG
Mflv_2230|M.gilvum_PYR-GCK DPVTRTAVVQPGLLNAEVKKVVAEHGLWYPPDPSSFEICSIGGNIATNAG
* : * .* : ::...: .* : * : .::** :****
Mb0166|M.bovis_AF2122/97 GLRTVRYGNMGEQVVGLDVALPDGTVLRRHSRVRRDNTGYDLPALFVGAE
Rv0161|M.tuberculosis_H37Rv GLRTVRYGNMGEQVVGLDVALPDGTVLRRHSRVRRDNTGYDLPALFVGAE
MMAR_0388|M.marinum_M GLRTVRYGNMGEQVVGLDVALPDGSVLRRHSLVRSDNTGYDLPALFVGAE
MUL_1038|M.ulcerans_Agy99 GLRTVRYGNMGEQVVGLDVALPDGSVLRRHSLVRSDNTGYDLPALFVGAE
MAV_5135|M.avium_104 GLHTVRYGNMGEQVLGIQVALPDGALLRRRSRVRRDNTGYDLPALFVGAE
MLBr_02631|M.leprae_Br4923 GLRTVRYGNMGEQVIGLEVALPDGSLLCRHSLARFDNTGYNLPALFVGAE
MSMEG_5045|M.smegmatis_MC2_155 GLRTVRYGNMGEQVLGLDIALPDGSVVHRHSQVRRDNTGYDLTSLFVGAE
Mvan_4473|M.vanbaalenii_PYR-1 GLRTVCYGNMGEQVIGLDVVLPDGSVVHRHSQVRSDNTGYDLASLFVGAE
TH_0754|M.thermoresistible__bu GLRTVRYGNMGEQVLGLEVALPDGSVLRRHSAVRRDNTGYDLTSLFIGAE
MAB_1398c|M.abscessus_ATCC_199 GLRTVRYGNMREQVIGIQVALPDGSIMERHSDVRADNTGYDLTSLFVGAE
Mflv_2230|M.gilvum_PYR-GCK GLCCVKYGVTTDYVLGLQVVLADGTAVRLGGPRLKDVAGLSLTKLFVGSE
** * ** : *:*:::.*.**: : . * :* .*. **:*:*
Mb0166|M.bovis_AF2122/97 GTLGVITALDLRLHPTPSHRVTAVCGFAELAALVDAGRMFRDVEGIAALE
Rv0161|M.tuberculosis_H37Rv GTLGVITALDLRLHPTPSHRVTAVCGFAELAALVDAGRMFRDVEGIAALE
MMAR_0388|M.marinum_M GTLGVITALDLRLHPTPSHRLTAVCGFADLEALVDAGRVFRDVGGIAALE
MUL_1038|M.ulcerans_Agy99 GTLGVITALDLRLHPTPSHRLTAVCGFADLEALVDAGRVFRDVGGIAALE
MAV_5135|M.avium_104 GTLGVITELDLRLHPTPTHTVTAICGFADLDAVVAAGRTFRDVDGIAALE
MLBr_02631|M.leprae_Br4923 GTLGVITALDLRLHPAPAHRVTAVCGFSDLGALVDASRAFRDVYGIAALE
MSMEG_5045|M.smegmatis_MC2_155 GTLGVITALDLRLYPVPTHRATAVCGFADLDALVETGRRFRDLDGIAALE
Mvan_4473|M.vanbaalenii_PYR-1 GTLGVITALDLRLHPTPRQRVTAICGFADLDALIETGRVFRDMEGIAALE
TH_0754|M.thermoresistible__bu GTLGVITALDLRLHPTPRHRVSAICGFTDLDALIDAGRVLRDTPGIAALE
MAB_1398c|M.abscessus_ATCC_199 GTLGVITALELRLHPVPTHSVAALTGFDSLDRLVEASRIFRDLDGIAALE
Mflv_2230|M.gilvum_PYR-GCK GTLGVVTEVTLKLLPAQHPACTVVATFDSVESAADAVVEITGRIRPSMLE
*****:* : *:* *. :.: * .: : : . : **
Mb0166|M.bovis_AF2122/97 LIDGRAAALTREHLGVRPPVEADWLLLVELAA----DHDQTDRLADLLGG
Rv0161|M.tuberculosis_H37Rv LIDGRAAALTREHLGVRPPVEADWLLLVELAA----DHDQTDRLADLLGG
MMAR_0388|M.marinum_M LIDGRAAALTREHLGVPAPVPGDWLLLVELAA----DHDQTERLADLLDG
MUL_1038|M.ulcerans_Agy99 LIDGRAAALTREHLGVPAPVPGDWLLLVELAA----DHDQTERLADLLDG
MAV_5135|M.avium_104 LIDGRAAALTREHRGVPTPVDADWLLLVELAA----DHDQTERLAGLLDD
MLBr_02631|M.leprae_Br4923 LIDGRASSLIREHLGVSSPVDGDWLLLVELAA----DHDQTNRLAELSMA
MSMEG_5045|M.smegmatis_MC2_155 LIDARAGRLAAEHLGVPVPVAGSWLLLIELAG----DTDQTERLADALTD
Mvan_4473|M.vanbaalenii_PYR-1 LIDARASALTAEHAGVAAPVQGAWQLLIELAG----ETDLTDRLAEALES
TH_0754|M.thermoresistible__bu LIDARIARLAAEHLGVAVPAPGDWLLLVELTSLGDADDDQTERLAVALDQ
MAB_1398c|M.abscessus_ATCC_199 LMDSR--------LGVPSPLDNPWLLLIELAR----DSDPTDDLAEALEA
Mflv_2230|M.gilvum_PYR-GCK FMDSTAINAVEDKLKMGLDRDAAAMMVAASDDRGAAGAQDAEYMARVFTE
::*. : :: : :: :*
Mb0166|M.bovis_AF2122/97 ARMCGEPAVGVDAAAQQRLWRTRESLAEVLGVYGPPLKFDVSLPLSAISG
Rv0161|M.tuberculosis_H37Rv ARMCGEPAVGVDAAAQQRLWRTRESLAEVLGVYGPPLKFDVSLPLSAISG
MMAR_0388|M.marinum_M VRLCGDPAVGVDVAAQQRLWRIRESLAEVLGVYGPPLKFDVSLPLAAISE
MUL_1038|M.ulcerans_Agy99 VRLCGDPAVGVDVAAQQRLWRIRESLAEVLGVYGPPLKFDVSLPLAAISE
MAV_5135|M.avium_104 APLSGEPAVGVDAVARQRLWQVREALAEVLGVYGPPLKFDVSLPLSSISE
MLBr_02631|M.leprae_Br4923 VQMCGVPAVGVDAGAQQRLWRVRESLAEVLGVYGPPLKFDVSLPLSAISG
MSMEG_5045|M.smegmatis_MC2_155 AQVCDEPAVGVDIGAQQRLWNVRESLAEVVGLFGPPLKFDVSLPLSAIGG
Mvan_4473|M.vanbaalenii_PYR-1 AELTDEPAVGVDANAQQRLWQVREAVAEVLGVYGPPLKFDVSLPLSAIRG
TH_0754|M.thermoresistible__bu LDLAGEPAVGVDSAARQRLWRVRESTAEVLGVHGPPVKFDVSLPPAAIPR
MAB_1398c|M.abscessus_ATCC_199 AGVAEHAAVGLDSTSRERLWQVRESVAEALGTYGPPLKFDVALPLAHIGE
Mflv_2230|M.gilvum_PYR-GCK HGATEVFST-SDPDEGDAFVAARRFAIPAVESKGSLLLEDVGVPLPALAE
:. * : : *. .: *. : **.:* . :
Mb0166|M.bovis_AF2122/97 FARDAVALVHRHVPDSPEALPLLFGHIGEGNLHLNVLRCPP-----DREP
Rv0161|M.tuberculosis_H37Rv FARDAVALVHRHVPDSPEALPLLFGHIGEGNLHLNVLRCPP-----DREP
MMAR_0388|M.marinum_M FAAAAATLVAAHVP---EAIPLLFGHIGEGNLHLNVLRCPV-----PREP
MUL_1038|M.ulcerans_Agy99 FAAAAATLVAAHVP---EAIPLLFGHIGEGNLHLNVLRCPV-----PREP
MAV_5135|M.avium_104 FERAAIDLVHTHAP---DALPVLFGHIGEGNLHLNVLRCAA-----DREH
MLBr_02631|M.leprae_Br4923 FAKEAATLIDAQVSDVQEALPVLFGHVGEGNLHLNVLRCPP-----DREK
MSMEG_5045|M.smegmatis_MC2_155 FAESAGKLVAAHAP---DAIPVLFGHIGEGNLHLNVLRCAL-----DTEP
Mvan_4473|M.vanbaalenii_PYR-1 FAEDAAALVAEHAP---DAIPVLFGHVGEGNLHLNILRCDL-----TGER
TH_0754|M.thermoresistible__bu FAAEAAAIVGRHAA---QAIPVLFGHIGEGNLHLNVLRCTP-----DEER
MAB_1398c|M.abscessus_ATCC_199 FERRASELIAAAVP---GAMPVLFGHVGEGNLHLNVLRCP-------DST
Mflv_2230|M.gilvum_PYR-GCK LVGGVEKIAAARDV-----MISVIAHAGDGNTHPLIVFDPSDTAMEQRAQ
: . : : ::.* *:** * ::
Mb0166|M.bovis_AF2122/97 ALYAKMMGLIAECGGNVSSEHGVGSRKRAYLGMSRQANDVAAMRRVKAAL
Rv0161|M.tuberculosis_H37Rv ALYAKMMGLIAECGGNVSSEHGVGSRKRAYLGMSRQANDVAAMRRVKAAL
MMAR_0388|M.marinum_M ALYQEMMSLIAQCGGNVSSEHGIGSRKRPYLQMSREDTDIAVMRAVKAAL
MUL_1038|M.ulcerans_Agy99 ALYQEMMSLIAQCGGNVSPEHGIGSRKRPYLQMSREDTDIAVMRAVKAAL
MAV_5135|M.avium_104 ALYEPMMDLIAASGGNVSSEHGVGSRKRPYLGMSREPADIAAMRTVKAAL
MLBr_02631|M.leprae_Br4923 TLTAAMMELIAQAGGNVSSEHGVGSNKRAYLRMSRDPADIAAMRTIKAAL
MSMEG_5045|M.smegmatis_MC2_155 ALYAAMMALIADCGGNVSSEHGVGTRKRDYVAMSRTPADIAAMRAVKAAL
Mvan_4473|M.vanbaalenii_PYR-1 GLYSAMMALIARHGGNVSSEHGVGTRKRDYLSMARTEADIAAMRTVKAAF
TH_0754|M.thermoresistible__bu ALYPAMMDLIAGCGGNVSSEHGVGTRKRDYLGMSRSDADIAAMRAIKAAF
MAB_1398c|M.abscessus_ATCC_199 TLYQPMMTLIADLGGNVSSEHGVGTLKRDYLGMARTAGDIAAMRAVKSAF
Mflv_2230|M.gilvum_PYR-GCK QAFGEIMDLAVSLGGTITGEHGVGRLKRPWLAGQLGPEAMELNRRIKQAL
:* * . **.:: ***:* ** :: : * :* *:
Mb0166|M.bovis_AF2122/97 DPTGYLNAAVLFD---
Rv0161|M.tuberculosis_H37Rv DPTGYLNAAVLFD---
MMAR_0388|M.marinum_M DPTGYLNPAVLFD---
MUL_1038|M.ulcerans_Agy99 DPTGYLNPAVLFD---
MAV_5135|M.avium_104 DPTGYLNAAVLFD---
MLBr_02631|M.leprae_Br4923 DPTGYLNAAVLFD---
MSMEG_5045|M.smegmatis_MC2_155 DPRGYLNPAVLFAQ--
Mvan_4473|M.vanbaalenii_PYR-1 DPTGYLNRAVLFG---
TH_0754|M.thermoresistible__bu DPTGYLNAAVLFG---
MAB_1398c|M.abscessus_ATCC_199 DPTGFLNPAVLFGPES
Mflv_2230|M.gilvum_PYR-GCK DPDGILNPGAAI----
** * ** .. :