For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVVAGINFGDAAFAATVQDRVVRIEQLMDAELRDTDEVMTESVLHLFAAGGKRFRPLFTVLSAQIGPRPD APEVIIAGAVIELVHLASLYHDDVMDEAEVRRGAPSANLRWSNSVAILAGDYLFATASRLVSQLGLDAVR IIADTFAQLVTGQMRETRGVPEGADSIEQYLKVVYEKTGCLIGSAGRLGGLFAGAHAEQVERLSRLGSIV GTAFQISDDIIDIDSDYTKSGKLPGTDVREGVRTLPILYALRETGPDSERLRALLAGPVDNDAEVVEALA LLRASPGMARARQVLAHYADQASGELAQLPDGPSRDALATLVDYTTNRHG
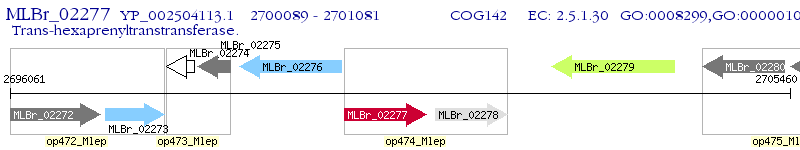
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02277 | - | - | 100% (330) | polyprenyl diphosphate synthase component |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0577 | grcC1 | 1e-141 | 77.20% (329) | polyprenyl-diphosphate synthase |
| M. gilvum PYR-GCK | Mflv_5253 | - | 1e-131 | 70.52% (329) | trans-hexaprenyltranstransferase |
| M. tuberculosis H37Rv | Rv0562 | grcC1 | 1e-141 | 77.20% (329) | polyprenyl-diphosphate synthase |
| M. abscessus ATCC 19977 | MAB_3928c | - | 1e-125 | 67.68% (328) | polyprenyl-diphosphate synthase GrcC1 |
| M. marinum M | MMAR_0911 | grcC1 | 1e-143 | 77.20% (329) | polyprenyl diphosphate synthetase, GrcC1 |
| M. avium 104 | MAV_4581 | - | 1e-137 | 76.00% (325) | polyprenyl synthetase |
| M. smegmatis MC2 155 | MSMEG_1133 | - | 1e-133 | 71.43% (329) | bifunctional short chain isoprenyl diphosphate synthase |
| M. thermoresistible (build 8) | TH_1767 | - | 1e-134 | 71.73% (329) | bifunctional short chain isoprenyl diphosphate synthase |
| M. ulcerans Agy99 | MUL_0664 | grcC1 | 1e-143 | 77.20% (329) | polyprenyl diphosphate synthetase, GrcC1 |
| M. vanbaalenii PYR-1 | Mvan_0992 | - | 1e-132 | 71.73% (329) | trans-hexaprenyltranstransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5253|M.gilvum_PYR-GCK MRTPANVVAGVDFGDAQFAQDVRDGVARIEALMSEELGRADALMSEAVNH
Mvan_0992|M.vanbaalenii_PYR-1 ------MVAGVDFGDAEFAQDVRDGVARIEALMSEELGKADVLMAEAVQH
MSMEG_1133|M.smegmatis_MC2_155 ------MVAGVDFGDPAFAADVRDGVARIEDLMSDELGKADELMAEAVQH
TH_1767|M.thermoresistible__bu ------VVAGVDLGDPDFAAGVRDGVARIEDLMSSELGKADDLMAEAVQH
Mb0577|M.bovis_AF2122/97 MRTPATVVAGVDLGDAVFAAAVRAGVARVEQLMDTELRQADEVMSDSLLH
Rv0562|M.tuberculosis_H37Rv MRTPATVVAGVDLGDAVFAAAVRAGVARVEQLMDTELRQADEVMSDSLLH
MMAR_0911|M.marinum_M MRTSATVVAGVDVGDAEFATTVRDGVASIERLMDAELRGADEVMTDSLLH
MUL_0664|M.ulcerans_Agy99 MRTSATVVAGVDVGDAEFATTVRDGVASIERLMDAELRGADEVMTDSLLH
MLBr_02277|M.leprae_Br4923 -----MVVAGINFGDAAFAATVQDRVVRIEQLMDAELRDTDEVMTESVLH
MAV_4581|M.avium_104 ----------MDFGDPAFAATVRDGVGRIEQLMDTELRSADDIMTELLTH
MAB_3928c|M.abscessus_ATCC_199 -------MAGVDLGDPVFAAKVRDHLAAIETLIVSELGQSDPLLTESVLH
::.**. ** *: : :* *: ** :* :::: : *
Mflv_5253|M.gilvum_PYR-GCK LFQAGGKRFRPLFTVLSAQLGPEPDAWQVTVAGAVIELVHLATLYHDDVM
Mvan_0992|M.vanbaalenii_PYR-1 LFQAGGKRFRPLFTVLSAQLGPEPDAWQVTVAGAVIELVHLATLYHDDVM
MSMEG_1133|M.smegmatis_MC2_155 LFQAGGKRFRPLFTVLSAQLGPEPDAWQVTVAGAVIELVHLATLYHDDVM
TH_1767|M.thermoresistible__bu LFRAGGKRFRPLFTVLSAQLGPEPDAWQVTVAGAVIELVHLATLYHDDVM
Mb0577|M.bovis_AF2122/97 LFNAGGKRFRPLFTVLSAQIGPQPDAAAVTVAGAVIEMIHLATLYHDDVM
Rv0562|M.tuberculosis_H37Rv LFNAGGKRFRPLFTVLSAQIGPQPDAAAVTVAGAVIEMIHLATLYHDDVM
MMAR_0911|M.marinum_M LFLAGGKRFRPLFTVLSAQVGPNPGADEVVIAGAVIEMIHLATLYHDDVM
MUL_0664|M.ulcerans_Agy99 LFLAGGKRFRPLFTVLSAQVGPNPGADEVVIAGAVIEMIHLATLYHDDVM
MLBr_02277|M.leprae_Br4923 LFAAGGKRFRPLFTVLSAQIGPRPDAPEVIIAGAVIELVHLASLYHDDVM
MAV_4581|M.avium_104 LFKAGGKRFRPLFTVLSAQIGPDPDAAEVTIAGAVIELVHLATLYHDDVM
MAB_3928c|M.abscessus_ATCC_199 LFHAGGKRFRPLFTVLAAQTGPDPDNDEVILAGAVCELIHLATLYHDDVM
** *************:** ** *. * :**** *::***:*******
Mflv_5253|M.gilvum_PYR-GCK DEAQVRRGAPSANARWGNNIAILAGDYLFATASRLTSRLGPEAVRIIADT
Mvan_0992|M.vanbaalenii_PYR-1 DEAQVRRGAPSANARWSNNIAILAGDYLFATASRLVSRLGPDAVRIIADT
MSMEG_1133|M.smegmatis_MC2_155 DEAQMRRGAQSANARWGNNIAILAGDYLFATASRLVSRLGPDAVRVIADT
TH_1767|M.thermoresistible__bu DDAEMRRGAPSANAKWGNSIAILAGDYLFATASRLVSRLGPEAVRVIADT
Mb0577|M.bovis_AF2122/97 DEAQVRRGAPSANAQWGNNVAILAGDYLLATASRLVARLGPEAVRIIADT
Rv0562|M.tuberculosis_H37Rv DEAQVRRGAPSANAQWGNNVAILAGDYLLATASRLVARLGPEAVRIIADT
MMAR_0911|M.marinum_M DEAQVRRGAPSANARWGNNVAILAGDYLLATASRLVSRLGPDAVRIVADT
MUL_0664|M.ulcerans_Agy99 DEAQVRRGAPSANARWGNNVAILAGDYLLATASRLVSRLGPDAVRIVADT
MLBr_02277|M.leprae_Br4923 DEAEVRRGAPSANLRWSNSVAILAGDYLFATASRLVSQLGLDAVRIIADT
MAV_4581|M.avium_104 DEAEVRRGTPTANVRWGNNVAILAGDYLFATASRLVSRLGPEAVQLIAET
MAB_3928c|M.abscessus_ATCC_199 DEAQKRRGVPSANARWGNNVAILAGDYLLATASRLVSRLGPTAVRIVAET
*:*: ***. :** :*.*.:********:******.::** **:::*:*
Mflv_5253|M.gilvum_PYR-GCK FALLVTGQMRETRGAADQVDSVDHYLKVVYEKTACLISASGRFGATFSGS
Mvan_0992|M.vanbaalenii_PYR-1 FALLVTGQMRETRGAADHVDEVDHYLKVVYEKTACLIAASGRFGATFSGA
MSMEG_1133|M.smegmatis_MC2_155 FAQLVTGQMRETRGAADHVDSVDHYLKVVYEKTACLIAASGRFGATFSGA
TH_1767|M.thermoresistible__bu FAQLVTGQMRETRGAADGADPIEHYLKVVYEKTACLISASGRFGATFSGA
Mb0577|M.bovis_AF2122/97 FAQLVTGQMRETRGTSENVDSIEQYLKVVQEKTGSLIGAAGRLGGMFSGA
Rv0562|M.tuberculosis_H37Rv FAQLVTGQMRETRGTSENVDSIEQYLKVVQEKTGSLIGAAGRLGGMFSGA
MMAR_0911|M.marinum_M FAQLVTGQMRETRGAPEGVDMIEQYLKVVHEKTGCLIGAAGRFGGMFSGA
MUL_0664|M.ulcerans_Agy99 FAQLVTGQMRETRGAPEGVDMIEQYLKVVHEKTGCLIGAAGRFGGMFSGA
MLBr_02277|M.leprae_Br4923 FAQLVTGQMRETRGVPEGADSIEQYLKVVYEKTGCLIGSAGRLGGLFAGA
MAV_4581|M.avium_104 FAQLVTGQMRETRGRKEGSDPIEHYLKVVYEKTACLIAAAGRFGAMFSGA
MAB_3928c|M.abscessus_ATCC_199 FAELVTGQMRETVGVPEGGDETEHYLKVIHEKTGSLIAAAGRFGAMTSRC
** ********* * : * ::****: ***..**.::**:*. : .
Mflv_5253|M.gilvum_PYR-GCK DEAQIERLAKLGGIVGTAFQISDDIIDIDSDADESGKVPGTDLREGVHTL
Mvan_0992|M.vanbaalenii_PYR-1 DDEQIERLARLGGIVGTAFQISDDIIDIDSDPDESGKVPGTDLREGVHTL
MSMEG_1133|M.smegmatis_MC2_155 NDEQIERLSRLGGIVGTAFQIADDIIDIDSDPDESGKTPGTDLREGVHTL
TH_1767|M.thermoresistible__bu DEEQVERLARLGGIVGTAFQISDDIIDIESESTESGKTPGTDLREGVHTL
Mb0577|M.bovis_AF2122/97 TDEQVERLSRLGGVVGTAFQIADDIIDIDSESDESGKLPGTDVREGVHTL
Rv0562|M.tuberculosis_H37Rv TDEQVERLSRLGGVVGTAFQIADDIIDIDSESDESGKLPGTDVREGVHTL
MMAR_0911|M.marinum_M DGEQVERLNRLGGIVGTAFQIADDIIDIDSDTEQSGKLPGTDLREGVRTL
MUL_0664|M.ulcerans_Agy99 DGEQVERLNRLGGIVGTAFQIADDIIDIDSDTEQSGKLPGTDLREGVRTL
MLBr_02277|M.leprae_Br4923 HAEQVERLSRLGSIVGTAFQISDDIIDIDSDYTKSGKLPGTDVREGVRTL
MAV_4581|M.avium_104 DADQVERLSRLGGIVGTAFQISDDIIDIDSDSHESGKLPGTDVREGVHTL
MAB_3928c|M.abscessus_ATCC_199 DDDQIERLSRLGAIVGTAFQISDDIIDIESLTDESGKTPGTDLREGVHTL
*:*** :**.:*******:******:* :*** ****:****:**
Mflv_5253|M.gilvum_PYR-GCK PVLYALRETGPDADRLRELLAGPVTDDADVAEALRLLRGSQGIRKAKETV
Mvan_0992|M.vanbaalenii_PYR-1 PVLYALRETGPDADRLRELLATPVTDDGDLAEALRLLRASDGITKAKETV
MSMEG_1133|M.smegmatis_MC2_155 PVLYALRETGPDADRLRELLAGPIERDEDVAEALALLRRSPGLTKAKETV
TH_1767|M.thermoresistible__bu PVLYALRETGPEAERLRELVSRPVEDDADLAEALDLLRRSPGMAQATETV
Mb0577|M.bovis_AF2122/97 PMLYALRESGPDCARLRALLNGPVDDDAEVREALTLLRASPGMARAKDVL
Rv0562|M.tuberculosis_H37Rv PMLYALRESGPDCARLRALLNGPVDDDAEVREALTLLRASPGMARAKDVL
MMAR_0911|M.marinum_M PMLYALRQTGSDGARLRTLLAGPIRDDAEVFEALALLRASPGMDQAKEVL
MUL_0664|M.ulcerans_Agy99 PMLYALRQTGSDGARLRTLLAGPIRDDAEVFEALALLRASPGMDQAKEVL
MLBr_02277|M.leprae_Br4923 PILYALRETGPDSERLRALLAGPVDNDAEVVEALALLRASPGMARARQVL
MAV_4581|M.avium_104 PMVYALREPGPDAARLRELLVGPIDDDEVVAEALTLLRASPGMAKAKQSL
MAB_3928c|M.abscessus_ATCC_199 PMLYALRDTGPDADRLRVLLAAPITDDAEVEEALALLRASEGLAQAKSVV
*::****:.*.: *** *: *: * : *** *** * *: :* . :
Mflv_5253|M.gilvum_PYR-GCK IGYAQEAQRELDQLPDVPGRQALASLVDYTVNRHG
Mvan_0992|M.vanbaalenii_PYR-1 AAYAREAEQELGQLPAGPGRQALATLVQYTINRHG
MSMEG_1133|M.smegmatis_MC2_155 ASYAAQAREELSHLPECPGRQALAVLVDYTISREG
TH_1767|M.thermoresistible__bu REYATEARSVLAQLPAGPGRDALATLIDFTVNRHG
Mb0577|M.bovis_AF2122/97 AQYAAQARHELALLPDVPGRRALAALVDYTVSRHG
Rv0562|M.tuberculosis_H37Rv AQYAAQARHELALLPDVPGRRALAALVDYTVSRHG
MMAR_0911|M.marinum_M GQYAARARHELDLLPDVPGRQALAALVDYTVNRHG
MUL_0664|M.ulcerans_Agy99 GQYAARARHELDLLPDVPGRQALAALVDYTVNRHG
MLBr_02277|M.leprae_Br4923 AHYADQASGELAQLPDGPSRDALATLVDYTTNRHG
MAV_4581|M.avium_104 REYAAQAHRELDQLPDVPGRRALQTLVDFTISRHG
MAB_3928c|M.abscessus_ATCC_199 IDYADRAKAELVELPPGDARDALGVLIEYTVRRHG
** .* * ** .* ** *:::* *.*