For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSAFISSLRTVDLRRKILFTLGILVLYRVSAALPSPGVNYRHVQQCIQEATAGEAGEIYSLINLFSGGAL LKLTVGVMPYITASIIVQLLTVVIPRFEELRKEGQAGQAKMTQYTRYLAIALAVLQATSIVALAANGGLL QGCQEDIISDQSIFSLVVIVLVMTGGAALVMWMGELITERGIGNGMSLLIFVGIAARIPAEGKQILDSRG GVIFAAVCLAALVIIVGVVFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLIYI PHLITQLVRSGSGGVGKSWWDKFVGTYLSDPADPVYINIYFGLIIFFTYFYVSVTFNPDERADEMKKFGG FIPGIRPGRPTADYLRYVLSRITLPGSIYLGAIAVLPNLFLQIGNGGEVQNLPFGGTAVLIMIGVGLDTV KQIESQLMQRNYEGFLK*
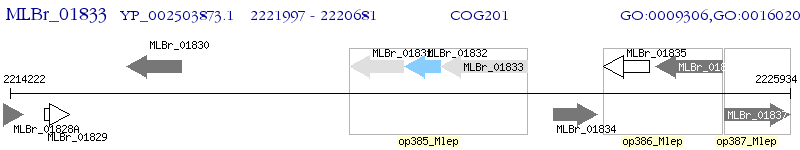
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01833 | secY | - | 100% (438) | preprotein translocase subunit SecY |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0753 | secY | 0.0 | 91.84% (441) | preprotein translocase subunit SecY |
| M. gilvum PYR-GCK | Mflv_5022 | secY | 0.0 | 80.67% (445) | preprotein translocase subunit SecY |
| M. tuberculosis H37Rv | Rv0732 | secY | 0.0 | 91.84% (441) | preprotein translocase subunit SecY |
| M. abscessus ATCC 19977 | MAB_3784c | secY | 0.0 | 79.95% (419) | preprotein translocase subunit SecY |
| M. marinum M | MMAR_1070 | secY | 0.0 | 91.38% (441) | preprotein translocase, SecY |
| M. avium 104 | MAV_4434 | secY | 0.0 | 92.29% (441) | preprotein translocase subunit SecY |
| M. smegmatis MC2 155 | MSMEG_1483 | secY | 0.0 | 85.71% (441) | preprotein translocase subunit SecY |
| M. thermoresistible (build 8) | TH_0325 | secY | 0.0 | 83.52% (443) | PROBABLE PREPROTEIN TRANSLOCASE SECY |
| M. ulcerans Agy99 | MUL_0828 | secY | 0.0 | 91.38% (441) | preprotein translocase subunit SecY |
| M. vanbaalenii PYR-1 | Mvan_1347 | secY | 0.0 | 81.57% (445) | preprotein translocase subunit SecY |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5022|M.gilvum_PYR-GCK ------------MLSAFISSLRTVDLRRKILFTLGIVILYRVGASIPSPG
Mvan_1347|M.vanbaalenii_PYR-1 ------------MLSAFISSLRTVDLRRKILFTLGIVILYRVGAQIPSPG
MSMEG_1483|M.smegmatis_MC2_155 ------------MLSAFISSLRTADLRRKILFTLGLVILYRVGASIPSPG
TH_0325|M.thermoresistible__bu ------------VLSAFISSLRTADLRRKILFTLGIVLLYRVGASIPSPG
MLBr_01833|M.leprae_Br4923 ------------MLSAFISSLRTVDLRRKILFTLGILVLYRVSAALPSPG
MAV_4434|M.avium_104 MMQEIPAAQEEELLSAFISSLRTVDLRRKILFTLGIVVLYRVGAALPSPG
MMAR_1070|M.marinum_M ------------MLSAFISSLRTVDLRRKILFTLGIVVLYRLGAALPSPG
MUL_0828|M.ulcerans_Agy99 ------------MLSAFISSLRTVDLRRKILFTLGIVVLYRLGAALPSPG
Mb0753|M.bovis_AF2122/97 ------------MLSAFISSLRTVDLRRKILFTLGIVILYRVGAALPSPG
Rv0732|M.tuberculosis_H37Rv ------------MLSAFISSLRTVDLRRKILFTLGIVILYRVGAALPSPG
MAB_3784c|M.abscessus_ATCC_199 ------------------------------------MVLYRVGATLPSPG
::***:.* :****
Mflv_5022|M.gilvum_PYR-GCK VNYPNVQQCIADVSGGESGQIYSLINLFSGGALLQLSVFAVGIMPYITAS
Mvan_1347|M.vanbaalenii_PYR-1 VNYPNVQQCIAEVSGGESGQIYSLINLFSGGALLQLSVFAVGIMPYITAS
MSMEG_1483|M.smegmatis_MC2_155 VNYPNVQQCIAQVSGGDSAQIYSLINLFSGGALLQLTVFAVGVMPYITAS
TH_0325|M.thermoresistible__bu VNYPNVQRCIEQVSGGDSGQIYSLINLFSGGALLQLAVFAVGVMPYITAS
MLBr_01833|M.leprae_Br4923 VNYRHVQQCIQEATAGEAGEIYSLINLFSGGALLKLT---VGVMPYITAS
MAV_4434|M.avium_104 VNYPNVQQCIKEASGGAAGQIYSLINLFSGGALLKLTVFAVGVMPYITAS
MMAR_1070|M.marinum_M VDFPNVQQCIKEASSGEAGQIYSLINLFSGGALLKLTVFAVGVMPYITAS
MUL_0828|M.ulcerans_Agy99 VDFPNVQQCIKEASSGEAGQIYSLINLFSGGALLKLTVFAVGVMPYITAS
Mb0753|M.bovis_AF2122/97 VNFPNVQQCIKEASAGEAGQIYSLINLFSGGALLKLTVFAVGVMPYITAS
Rv0732|M.tuberculosis_H37Rv VNFPNVQQCIKEASAGEAGQIYSLINLFSGGALLKLTVFAVGVMPYITAS
MAB_3784c|M.abscessus_ATCC_199 VNYANVKYCVEQVSSGDSAQIYSLINLFSGGALLQLSVFAVGVMPYITAS
*:: :*: *: :.:.* :.:**************:*: **:*******
Mflv_5022|M.gilvum_PYR-GCK IIVQLLGVVIPRFEQLRKEGQAGQTKLTQYTRYLAIALAILQATSIVALA
Mvan_1347|M.vanbaalenii_PYR-1 IIVQLLGVVIPRFEQLRKEGQAGQTKLTQYTRYLAIALAILQATSIVALA
MSMEG_1483|M.smegmatis_MC2_155 IIVQLLTVVIPRFEQLRKEGQAGQAKMTQYTRYLSIALAILQATSIVALA
TH_0325|M.thermoresistible__bu IIVQLLTVVIPRFEQLRKEGQAGQAKMTQYTRYLAVALALLQATSIVALA
MLBr_01833|M.leprae_Br4923 IIVQLLTVVIPRFEELRKEGQAGQAKMTQYTRYLAIALAVLQATSIVALA
MAV_4434|M.avium_104 IIVQLLTVVIPRFEELRKEGQSGQAKMTQYTRYLAIALAILQATSIVALA
MMAR_1070|M.marinum_M IIVQLLTVVIPRFEELRKEGQSGQAKMTQYTRYLAIALAVLQATSIVALA
MUL_0828|M.ulcerans_Agy99 IIVQLLTVVIPRFEELRKEGQSGQAKMTQYTRYLAIALAVLQATSIVALA
Mb0753|M.bovis_AF2122/97 IIVQLLTVVIPRFEELRKEGQAGQSKMTQYTRYLAIALAILQATSIVALA
Rv0732|M.tuberculosis_H37Rv IIVQLLTVVIPRFEELRKEGQAGQSKMTQYTRYLAIALAILQATSIVALA
MAB_3784c|M.abscessus_ATCC_199 IIVQLLTVVIPRFEQLRKEGQSGQAKMTQYTRYLSVALALLQSTSIVALG
****** *******:******:**:*:*******::***:**:******.
Mflv_5022|M.gilvum_PYR-GCK ANGGLLQGCS--LDIIQGQSEGLNVWTLAVIVIVMTAGAALVMWMGELVT
Mvan_1347|M.vanbaalenii_PYR-1 ANGGLLQGCA--LDIIEGQSEGLNIWTLTVIVIVMTAGAALVMWMGELVT
MSMEG_1483|M.smegmatis_MC2_155 ANGGLLQGCS--LDIIQDSS----IFGLIVIVLVMTAGAALVMWMGELVT
TH_0325|M.thermoresistible__bu ANGGLLQGCT--LDIIDNSS----IFSLFVIVIVMTAGGALVMWMGELVT
MLBr_01833|M.leprae_Br4923 ANGGLLQGCQ--EDIISDQS----IFSLVVIVLVMTGGAALVMWMGELIT
MAV_4434|M.avium_104 ANGGLLQGCS--LDIIADQS----IFTLVVIVLVMTAGAALVMWMGELIT
MMAR_1070|M.marinum_M ANGGLLQGCS--LEIIADQS----IFTLVVIVMVMTAGAALVMWMGELIT
MUL_0828|M.ulcerans_Agy99 ANGGLLQGCS--LEIIADQS----IFTLVVIVMVMTAGAALVMWMGELIT
Mb0753|M.bovis_AF2122/97 ANGGLLQGCS--LDIIADQS----IFTLVVIVLVMTGGAALVMWMGELIT
Rv0732|M.tuberculosis_H37Rv ANGGLLQGCS--LDIIADQS----IFTLVVIVLVMTGGAALVMWMGELIT
MAB_3784c|M.abscessus_ATCC_199 ASGNLLQGCDRKDEIVADQS----IFALSVMVLVMTAGSTLVMWLGEMVT
*.*.***** :*: ..* :: * *:*:***.*.:****:**::*
Mflv_5022|M.gilvum_PYR-GCK ERGVGNGMSLIIFASIASAIPGEGKNILDSRGGMVFTLVCVAALIIVVGV
Mvan_1347|M.vanbaalenii_PYR-1 ERGVGNGMSLIIFASIAASIPGEGKNILDSRGGMVFTLVCVAAVVIVVGV
MSMEG_1483|M.smegmatis_MC2_155 ERGIGNGMSLLIFAGIAARIPAEGQTILESRGGVVFTAVVIAALVIIIGV
TH_0325|M.thermoresistible__bu ERGIGNGMSLLIFAGIAAQIPAEGNMILESRGGLVFTAVCAAALVILIGV
MLBr_01833|M.leprae_Br4923 ERGIGNGMSLLIFVGIAARIPAEGKQILDSRGGVIFAAVCLAALVIIVGV
MAV_4434|M.avium_104 ERGIGNGMSLLIFVGIAARIPAEGKTILDSRGGMIFAAVLVAALIIIVGV
MMAR_1070|M.marinum_M ERGIGNGMSLLIFVGIAARIPAEGNQIMESRGGVIFAAVCAATLIIIVGV
MUL_0828|M.ulcerans_Agy99 ERGIGNGMSLLIFVGIAARIPAEGNQIMESRGGVIFAAVCAATLIIIVGV
Mb0753|M.bovis_AF2122/97 ERGIGNGMSLLIFVGIAARIPAEGQSILESRGGVVFTAVCAAALIIIVGV
Rv0732|M.tuberculosis_H37Rv ERGIGNGMSLLIFVGIAARIPAEGQSILESRGGVVFTAVCAAALIIIVGV
MAB_3784c|M.abscessus_ATCC_199 ERGIGNGMSLLIFAGIAARIPAEGKTILDSRGGVTFAAVCVAALLIVVGV
***:******:**..**: **.**: *::****: *: * *:::*::**
Mflv_5022|M.gilvum_PYR-GCK VFVEQGQRRIPVQYAKRMVGRKMYGGTSTYLPLKVNQAGVIPVIFASSLI
Mvan_1347|M.vanbaalenii_PYR-1 VFVEQGQRRIPVQYAKRMVGRKMYGGTSTYLPLKVNQAGVIPVIFASSLI
MSMEG_1483|M.smegmatis_MC2_155 VFVEQGQRRIPVQYAKRMVGRKMYGGTSTYLPLKVNQAGVIPVIFASSLI
TH_0325|M.thermoresistible__bu VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
MLBr_01833|M.leprae_Br4923 VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
MAV_4434|M.avium_104 VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
MMAR_1070|M.marinum_M VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
MUL_0828|M.ulcerans_Agy99 VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
Mb0753|M.bovis_AF2122/97 VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
Rv0732|M.tuberculosis_H37Rv VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLI
MAB_3784c|M.abscessus_ATCC_199 VFVEQGQRRIPVQYAKRMVGRRMYGGTSTYLPLKVNQAGVIPVIFASSLL
*********************:***************************:
Mflv_5022|M.gilvum_PYR-GCK YIPHLITQLITSA-SDSPSTGWWQSFVADYLTDPSSPVYIAIYFGLIIFF
Mvan_1347|M.vanbaalenii_PYR-1 YIPHLITQLITSA-SDTPSTGWWQTFVAEYLTDPSSPVYIAIYFGLIVFF
MSMEG_1483|M.smegmatis_MC2_155 YIPHLITQLIQSG-SSNPGTGWWDKFVADYLTNPADPVYIAVYFGLIVFF
TH_0325|M.thermoresistible__bu YIPQLITQLIQSGRSDPTQTGWWDRFVAEYLTDPSSPVYIGIYFGLIIFF
MLBr_01833|M.leprae_Br4923 YIPHLITQLVRSG-SGGVGKSWWDKFVGTYLSDPADPVYINIYFGLIIFF
MAV_4434|M.avium_104 YIPHLITQLIRSG-SGGVGNSWWDKFVGSYLSDPSDPVYIGIYFGLIIFF
MMAR_1070|M.marinum_M YIPHLITQLVRSG-SGGVGHSWWDKFVGTYLSDPSNLGYIAIYFGLIIFF
MUL_0828|M.ulcerans_Agy99 YIPHLITQLVRSG-SGGVGHSWWDKFVGTYLSDPSNLGYIAIYFGLIIFF
Mb0753|M.bovis_AF2122/97 YIPHLITQLIRSG-SGVVGNSWWDKFVGTYLSDPSNLVYIGIYFGLIIFF
Rv0732|M.tuberculosis_H37Rv YIPHLITQLIRSG-SGVVGNSWWDKFVGTYLSDPSNLVYIGIYFGLIIFF
MAB_3784c|M.abscessus_ATCC_199 YIPGLITQLVRSG-GNKEKTGWWDRFVSTYLTDPSNYWYIAIYFTLIVFF
*** *****: *. .. .**: **. **::*:. ** :** **:**
Mflv_5022|M.gilvum_PYR-GCK TYFYVSITFNPEERADEMKKYGGFIPGIRPGKPTAQYLSFVLSRITLPGS
Mvan_1347|M.vanbaalenii_PYR-1 TYFYVSITFNPDERADEMKKYGGFIPGIRPGRPTADYLRFVLSRITLPGS
MSMEG_1483|M.smegmatis_MC2_155 TYFYVSITFNPDERADEMKKFGGFIPGIRPGKPTADYLRYVLSRITLPGS
TH_0325|M.thermoresistible__bu TYFYVSITFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
MLBr_01833|M.leprae_Br4923 TYFYVSVTFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
MAV_4434|M.avium_104 TYFYVSITFNPDERADEMKKFGGFIPGIRPGKPTADYLRYVLSRITLPGS
MMAR_1070|M.marinum_M TYFYVSITFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
MUL_0828|M.ulcerans_Agy99 TYFYVSITFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
Mb0753|M.bovis_AF2122/97 TYFYVSITFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
Rv0732|M.tuberculosis_H37Rv TYFYVSITFNPDERADEMKKFGGFIPGIRPGRPTADYLRYVLSRITLPGS
MAB_3784c|M.abscessus_ATCC_199 TFFYVSITFNPDERADEMNKFGGFIPGIRPGRATADYLRYVLNRITWPGS
*:****:****:******:*:**********:.**:** :**.*** ***
Mflv_5022|M.gilvum_PYR-GCK IYLGVIAVLPNLFLEIGNTG-SVQNLPFGGTAVLIAVGVGLDTVKQIESQ
Mvan_1347|M.vanbaalenii_PYR-1 IYLGIIAVLPNLFLEIGNTG-SVQNLPFGGTAVLIAVGVALDTIKQIESQ
MSMEG_1483|M.smegmatis_MC2_155 IYLGVIAVLPNLFLEIGNTG-SVQNLPFGGTAVLIMIGVGLDTVKQIESQ
TH_0325|M.thermoresistible__bu IYLGVIAVMPNVFLQMGSGGQTLQNLPFGGVAVLIMIGVGLDTVKQIESQ
MLBr_01833|M.leprae_Br4923 IYLGAIAVLPNLFLQIGNGG-EVQNLPFGGTAVLIMIGVGLDTVKQIESQ
MAV_4434|M.avium_104 IYLGAISVLPNLFLQIGNGG-AVQNLPFGGTAVLIMIGVGLDTVKQIESQ
MMAR_1070|M.marinum_M IYLGVIAVLPNLFLQIGNSG-GVQNLPFGGTAVLIMIGVGLDTVKQIESQ
MUL_0828|M.ulcerans_Agy99 IYLGVIAVLPNLFLQIGNSG-GVQNLPFGGTAVLIMIGVGLDTVKQIESQ
Mb0753|M.bovis_AF2122/97 IYLGVIAVLPNLFLQIGAGG-TVQNLPFGGTAVLIMIGVGLDTVKQIESQ
Rv0732|M.tuberculosis_H37Rv IYLGVIAVLPNLFLQIGAGG-TVQNLPFGGTAVLIMIGVGLDTVKQIESQ
MAB_3784c|M.abscessus_ATCC_199 IYLGVIAILPNLFLQIGSSG-GAQNLPFGGTAVLIMIGVGLDTVKQIESQ
**** *:::**:**::* * *******.**** :**.***:******
Mflv_5022|M.gilvum_PYR-GCK LMQRNYEGFLK
Mvan_1347|M.vanbaalenii_PYR-1 LMQRNYEGFLK
MSMEG_1483|M.smegmatis_MC2_155 LMQRNYEGFLK
TH_0325|M.thermoresistible__bu LMQRNYEGFLK
MLBr_01833|M.leprae_Br4923 LMQRNYEGFLK
MAV_4434|M.avium_104 LMQRNYEGFLK
MMAR_1070|M.marinum_M LMQRNYEGFLK
MUL_0828|M.ulcerans_Agy99 LMQRNYEGFLK
Mb0753|M.bovis_AF2122/97 LMQRNYEGFLK
Rv0732|M.tuberculosis_H37Rv LMQRNYEGFLK
MAB_3784c|M.abscessus_ATCC_199 LMQRNYEGFLK
***********