For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IEADYAAWLKERWEHFEHHTPLRVVNCPDYHLIRVAHELIAQVLNEHLDSKVTVLLFRRKYSPFLCQLLH DRTTDKMVRAVGLIRDAIALIMLDDIESRISRLVNSEGSSSAE*
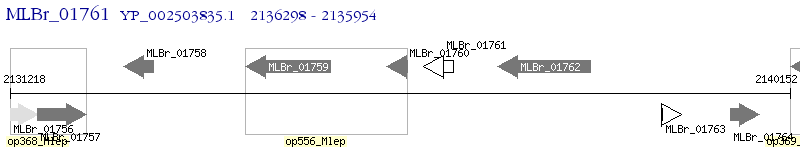
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01761 | - | - | 100% (114) | hypothetical protein MLBr_01761 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1318 | - | 3e-23 | 49.50% (101) | OB-fold nucleic acid binding domain-containing protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 MTSSTGNLRIPLSFGDVAKRVFLGKPLITEDIASEKLSNGVALGALSPDA
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 VSSVAYGPEQILIELLPHAGLAAFLLLLPITGVILLILVLVAASYRQVVM
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 AYTRAGGSYIVARDNFGPRVAQIAAAALLIDYVVTVAVQSAAGTVAVVSA
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 IPVLGPYSLAITVGVVLVICYANLRGLREAGMPFAVATYSFIVMVGLTIV
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 IGVVRAVIGNLPIYDPAHLAGTVPVHQGNGLVLGATILVLLRAFANGGSS
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 LTGVEAISNTVDYFRKPQGRNARRVLTAMASILGFLLAGVAYLAHVTHAT
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 PYLTEYPSVLSQIGHAVYGGGVVGDIFFVMVQASTAAILFTGANTSFNGF
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 PALASFVAEDRFLPRQLTKRGHRLVFSNGIITLTALSVALLVATGGSVNA
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 LVPFYAIGVFTGFAMAGYGMTRHHLTHREAGWRHRLTINLSAAVLSTIVV
MLBr_01761|M.leprae_Br4923 --------------------------------------------------
MAV_1318|M.avium_104 GIFAVAKFTEGAWLVVVVFPLLVFVLTRLNREYRAEAAILEMFRTDRPEL
MLBr_01761|M.leprae_Br4923 ---------------------------------------MIEADYAAWLK
MAV_1318|M.avium_104 VKYARHKVLVFVDSVDLAVIEALRYGRGLRADELVAVHFMVDPAHAAQLR
*::. :** *:
MLBr_01761|M.leprae_Br4923 ERWEHFEHHTPLRVVNCPDYHLIRVAHELIAQVLNEHLDSKVTVLLFRRK
MAV_1318|M.avium_104 NRWDHFSLDTPLRIVDCPDRRISRSAQLMVAKVRDEHPNTNVTVLLPRRT
:**:**. .****:*:*** :: * *: ::*:* :** :::***** **.
MLBr_01761|M.leprae_Br4923 YSPFLCQLLHDRTTDKMVRAVGLIRDAIALIMLDDIESRISR--------
MAV_1318|M.avium_104 FARLLGRLLHDRTADKVARAVSQIPDAAATIVPYDVESRIREAYPDTFEQ
:: :* :******:**:.***. * ** * *: *:**** .
MLBr_01761|M.leprae_Br4923 --------------------------------------LVNSE-------
MAV_1318|M.avium_104 RIAKELDKIQAWVSQDEDQKVAAYEHPARSSAVIMVAGLISGQRATIEGR
*:..:
MLBr_01761|M.leprae_Br4923 -------------------GSSSAE-------------------------
MAV_1318|M.avium_104 VSEVEDVAERGGTRRRIVVGDSSGEITVIFHHGHGGADIQPGQLLRISGK
*.**.*
MLBr_01761|M.leprae_Br4923 --------------------------------------------
MAV_1318|M.avium_104 ARQAGQRELSMVDPAYEVIEDPAREAEESGDSEESGDSQRADKK