For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FYDDNADLSIIQRRKVGVIGYGSQGYAHSLSLRDSGVQVRVGLPEGSKSRPKAFEQGLDVDTPAEVAKWA DVIMLLVPDTAQADVFENDIEPNLQPGDALFFGHGLNIHFELVKPSGDVTVAMVAPKGPGYLVRRQFVDG KGVPCLIAVNQDPTGTGEALALSYAKAIGGTRAGVIKTTFKDETETDLFGEQAVLCGGTEELVKAGFDVM VEAGYPPEMAYFEVLHELKLIVDLMYEGGIARMNYSVSDTAEFGGYLSGPRVIDADTKERMRGILRDIQD GSFVKKLVANVEGGNKQLEVLRKENAEHPIEVTGKKLRDLMSWVDRQSTETA*
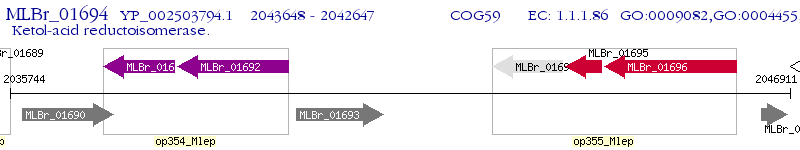
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01694 | ilvC | - | 100% (333) | ketol-acid reductoisomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3026c | ilvC | 1e-167 | 86.19% (333) | ketol-acid reductoisomerase |
| M. gilvum PYR-GCK | Mflv_4237 | - | 1e-165 | 85.59% (333) | ketol-acid reductoisomerase |
| M. tuberculosis H37Rv | Rv3001c | ilvC | 1e-167 | 86.19% (333) | ketol-acid reductoisomerase |
| M. abscessus ATCC 19977 | MAB_3321c | - | 1e-164 | 85.29% (333) | ketol-acid reductoisomerase |
| M. marinum M | MMAR_1712 | ilvC | 1e-173 | 89.79% (333) | ketol-acid reductoisomerase IlvC |
| M. avium 104 | MAV_3850 | ilvC | 1e-172 | 89.79% (333) | ketol-acid reductoisomerase |
| M. smegmatis MC2 155 | MSMEG_2374 | ilvC | 1e-165 | 85.59% (333) | ketol-acid reductoisomerase |
| M. thermoresistible (build 8) | TH_0064 | ilvC | 1e-165 | 85.59% (333) | PROBABLE KETOL-ACID REDUCTOISOMERASE ILVC (Acetohydroxy-acid |
| M. ulcerans Agy99 | MUL_1949 | ilvC | 1e-172 | 89.19% (333) | ketol-acid reductoisomerase |
| M. vanbaalenii PYR-1 | Mvan_2124 | - | 1e-165 | 85.29% (333) | ketol-acid reductoisomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3026c|M.bovis_AF2122/97 ----------MFYDDDADLSIIQGRKVGVIGYGSQGHAHSLSLRDSGVQV
Rv3001c|M.tuberculosis_H37Rv ----------MFYDDDADLSIIQGRKVGVIGYGSQGHAHSLSLRDSGVQV
MMAR_1712|M.marinum_M MADVSSQALEMFYDDDADLSIIQGRKVGVIGYGSQGHAHSLSLRDSGVQV
MUL_1949|M.ulcerans_Agy99 ----------MFYDDDADLSIIEGRKVGVIGYGSQGHAHSLSLRDSGVQV
MLBr_01694|M.leprae_Br4923 ----------MFYDDNADLSIIQRRKVGVIGYGSQGYAHSLSLRDSGVQV
MAV_3850|M.avium_104 ----------MFYDDDADLTIIQGRKVGVIGYGSQGHAHSLSLRDSGVQV
MSMEG_2374|M.smegmatis_MC2_155 ------MAVEMFYDDDADLSIIQGRKVAVIGYGSQGHAHSLSLRDSGVQV
Mvan_2124|M.vanbaalenii_PYR-1 ------MAVEMFYDADADLSIIQGRKVAVIGYGSQGHAHSLSLRDSGVQV
Mflv_4237|M.gilvum_PYR-GCK ------MAVEMFYDADADLSIIQGRKVAVIGYGSQGHAHSLSLRDSGVEV
MAB_3321c|M.abscessus_ATCC_199 ----------MFYDDDADLTLIQGRKVGVIGYGSQGHAHSLSLRDSGVQV
TH_0064|M.thermoresistible__bu ----------MFYDDDADLSIIQGRKVGVIGYGSQGHAHALSLRDSGVQV
**** :***::*: ***.********:**:********:*
Mb3026c|M.bovis_AF2122/97 RVGLKQGSRSRPKVEEQGLDVDTPAEVAKWADVVMVLAPDTAQAEIFAGD
Rv3001c|M.tuberculosis_H37Rv RVGLKQGSRSRPKVEEQGLDVDTPAEVAKWADVVMVLAPDTAQAEIFAGD
MMAR_1712|M.marinum_M RVGLKEGSKSRAKVEEQGIEVDTPAKVAEWADVIMVLAPDTAQAEIFAND
MUL_1949|M.ulcerans_Agy99 RVGLKEGSKSRAKVEEQGIEVDTPAKVAEWADVIMVLAPDTAQAEIFAND
MLBr_01694|M.leprae_Br4923 RVGLPEGSKSRPKAFEQGLDVDTPAEVAKWADVIMLLVPDTAQADVFEND
MAV_3850|M.avium_104 KVGLKEGSKSRAKVSEQGLDVDTPAAVAKWADVIMLLAPDTAQADIFKND
MSMEG_2374|M.smegmatis_MC2_155 KVGLKEGSKSREKAEEQGLEVDTPAEVAKWADVIMLLAPDTAQASIFTND
Mvan_2124|M.vanbaalenii_PYR-1 KVGLKEGSKSREKVTEQGLEVDTPAEVAKWADVIMLLAPDTAQAEIFTND
Mflv_4237|M.gilvum_PYR-GCK KVGLKEGSKSRDKVTEQGLDVDTPAEVAKWADVIMLLAPDTAQAEIFTKD
MAB_3321c|M.abscessus_ATCC_199 KVGLREGSKSREKVTEQGLEVDTPEEVAKWADVIMILAPDTAQAEIFKNE
TH_0064|M.thermoresistible__bu KVGLREGSKSREKVAEQGLEVDTPAEVAKWADVIMLLAPDTAQADIFKND
:*** :**:** *. ***::**** **:****:*:*.******.:* :
Mb3026c|M.bovis_AF2122/97 IEPNLKPGDALFFGHGLNVHFGLIKPPADVAVAMVAPKGPGHLVRRQFVD
Rv3001c|M.tuberculosis_H37Rv IEPNLKPGDALFFGHGLNVHFGLIKPPADVAVAMVAPKGPGHLVRRQFVD
MMAR_1712|M.marinum_M IEPNLKPGDALFFGHGLNVHFGLIKPPADVTVAMVAPKGPGHLVRRQFVD
MUL_1949|M.ulcerans_Agy99 IEPNLKPGDALFFGHGLNVHFGLIKPPADVTVAMVAPKGPGHLVRRQFVD
MLBr_01694|M.leprae_Br4923 IEPNLQPGDALFFGHGLNIHFELVKPSGDVTVAMVAPKGPGYLVRRQFVD
MAV_3850|M.avium_104 IEPNLSDGDALFFGHGLNIHFGLIKPPAEVTVAMVAPKGPGHLVRRQFVD
MSMEG_2374|M.smegmatis_MC2_155 IEPNLEDGNALFFGHGLNIHFGLIKAPENVTVGMVAPKGPGHLVRRQFVD
Mvan_2124|M.vanbaalenii_PYR-1 IEPNLEDGNALFFGHGLNIHFGLIKPPANVTIGMVAPKGPGHLVRRQFVD
Mflv_4237|M.gilvum_PYR-GCK IEPNLEDGNALFFGHGLNIHFGLIKPPANVTIGMVAPKGPGHLVRRQFVD
MAB_3321c|M.abscessus_ATCC_199 IEPHLKDGDALFFGHGLNIHFGLIKPPANVTVAMVAPKGPGHLVRRQFVD
TH_0064|M.thermoresistible__bu IEPHLNDGDALFFGHGLNIHFGLIQPPANVVVGMVAPKGPGHLVRRQFVD
***:*. *:*********:** *::.. :*.:.********:********
Mb3026c|M.bovis_AF2122/97 GKGVPCLVAVEQDPRGDGLALALSYAKAIGGTRAGVIKTTFKDETETDLF
Rv3001c|M.tuberculosis_H37Rv GKGVPCLVAVEQDPRGDGLALALSYAKAIGGTRAGVIKTTFKDETETDLF
MMAR_1712|M.marinum_M GKGVPCLIAVDQDPTGKGEALALSYAKAIGGTRAGVIKTTFKDETETDLF
MUL_1949|M.ulcerans_Agy99 GKGVPCLIAVDQDPTGKGEALALSYAKAIGGTRAGVIKTTFKDETETDLF
MLBr_01694|M.leprae_Br4923 GKGVPCLIAVNQDPTGTGEALALSYAKAIGGTRAGVIKTTFKDETETDLF
MAV_3850|M.avium_104 GKGVPCLIAVDQDPTGKGEALALSYAKAIGGTRAGVIKTTFKDETETDLF
MSMEG_2374|M.smegmatis_MC2_155 GKGVPCLIAIDQDPKGEGQALALSYAAAIGGARAGVIKTTFKEETETDLF
Mvan_2124|M.vanbaalenii_PYR-1 GKGVPCLIAIDQDPKGEGQALALSYAAAIGGARAGVIKTTFKEETETDLF
Mflv_4237|M.gilvum_PYR-GCK GKGVPCLIAVDQDPKGEGQALALSYAAAIGGARAGVIKTTFKDETETDLF
MAB_3321c|M.abscessus_ATCC_199 GKGVPALIAVHQDPKGEGQALALSYAKGIGGTRAGVIKTDFKEETETDLF
TH_0064|M.thermoresistible__bu GKGVPCLVAVQQDPKGNGLALALSYAKGIGGTRAGVIKTTFKDETETDLF
*****.*:*:.*** * * ******* .***:******* **:*******
Mb3026c|M.bovis_AF2122/97 GEQTVLCGGTEELVKAGFEVMVEAGYPAELAYFEVLHELKLIVDLMYEGG
Rv3001c|M.tuberculosis_H37Rv GEQTVLCGGTEELVKAGFEVMVEAGYPAELAYFEVLHELKLIVDLMYEGG
MMAR_1712|M.marinum_M GEQAVLCGGTEELVKTGFDVMVEAGYPPEMAYFEVLHELKLIVDLMYEGG
MUL_1949|M.ulcerans_Agy99 GEQAVLCGGTEELVKTGFDVMVEAGYPPEMAYFEVLHELKLIVDLMYEGG
MLBr_01694|M.leprae_Br4923 GEQAVLCGGTEELVKAGFDVMVEAGYPPEMAYFEVLHELKLIVDLMYEGG
MAV_3850|M.avium_104 GEQAVLCGGTEELVKAGFDVMVEAGYPPEMAYFEVLHELKLIVDLMYEGG
MSMEG_2374|M.smegmatis_MC2_155 GEQAVLCGGTEELIKTGFEVMVEAGYAPEMAYFEVLHELKLIVDLIYEGG
Mvan_2124|M.vanbaalenii_PYR-1 GEQAVLCGGTEELVKTGFEVMVEAGYAPEMAYFEVLHELKLIVDLMYEGG
Mflv_4237|M.gilvum_PYR-GCK GEQAVLCGGTEELVKTGFDVMVEAGYEPELAYFEVLHELKLIVDLMYEGG
MAB_3321c|M.abscessus_ATCC_199 GEQAVLCGGTEELVKTGFDVMVEAGYAPEMAYFEVLHELKLIVDLMYEGG
TH_0064|M.thermoresistible__bu GEQAVLCGGTEELIKTGFDVMVEAGYAPELAYFEVLHELKLIVDLIYEGG
***:*********:*:**:******* .*:***************:****
Mb3026c|M.bovis_AF2122/97 LARMYYSVSDTAEFGGYLSGPRVIDAGTKERMRDILREIQDGSFVHKLVA
Rv3001c|M.tuberculosis_H37Rv LARMYYSVSDTAEFGGYLSGPRVIDAGTKERMRDILREIQDGSFVHKLVA
MMAR_1712|M.marinum_M IARMNYSVSDTAEFGGYLSGPRVIDAGTKERMRAILRDIQSGDFVKKLVA
MUL_1949|M.ulcerans_Agy99 IARMNYSVSDTAEFGGYLSGPRVIDAGTKERMRAILRDIQSGDFVKKLVA
MLBr_01694|M.leprae_Br4923 IARMNYSVSDTAEFGGYLSGPRVIDADTKERMRGILRDIQDGSFVKKLVA
MAV_3850|M.avium_104 IARMNYSVSDTAEFGGYLSGPRVIDAGTKDRMREILRDIQNGDFVKKLVA
MSMEG_2374|M.smegmatis_MC2_155 IARMNYSVSDTAEFGGYLSGPRVIDADTKKRMQDILKDIQDGSFVKRLVA
Mvan_2124|M.vanbaalenii_PYR-1 IARMNYSVSDTAEFGGYLSGPRVIDADTKKRMQDILKDIQDGTFVKRLVA
Mflv_4237|M.gilvum_PYR-GCK IARMNYSVSDTAEFGGYLSGPRVIDADTKKRMQDILKDIQDGTFVKRLVA
MAB_3321c|M.abscessus_ATCC_199 IARMNYSVSDTAEFGGYLSGPRVIDAGTKDRMRQILKEIQDGTFVKRLVA
TH_0064|M.thermoresistible__bu IARMNYSVSDTAEFGGYLSGPRVIDAGTKERMKQILAEIQDGTFVKKLVA
:*** *********************.**.**: ** :**.* **::***
Mb3026c|M.bovis_AF2122/97 DVEGGNKQLEELRRQNAEHPIEVVGKKLRDLMSWVDRPITETA
Rv3001c|M.tuberculosis_H37Rv DVEGGNKQLEELRRQNAEHPIEVVGKKLRDLMSWVDRPITETA
MMAR_1712|M.marinum_M NVEGGNKQLEALRKENAEHPIEVTGKKLRDLMSWVDRPITETA
MUL_1949|M.ulcerans_Agy99 NVEGGNKQLEALRKENAEHSIEVTGKKLRDLMSWVDRPITETA
MLBr_01694|M.leprae_Br4923 NVEGGNKQLEVLRKENAEHPIEVTGKKLRDLMSWVDRQSTETA
MAV_3850|M.avium_104 NVEGGNKQLEQLRKENAEHPIEVVGKRLRDLMSWVDRPITETA
MSMEG_2374|M.smegmatis_MC2_155 NVEGGNKELEALRKANAEHPIEVTGKKLRDLMSWVDRPITETA
Mvan_2124|M.vanbaalenii_PYR-1 NVEGGNKELEALRKKNAEHPIEVTGKKLRDLMSWVDRPITETA
Mflv_4237|M.gilvum_PYR-GCK NVEGGNKELEGLRKQNAEHPIEVTGKKLRDLMSWVDRPITETA
MAB_3321c|M.abscessus_ATCC_199 NVEGGNKELEGLRKENAEHPIEVTGKKLRDLMSWVDRPITETA
TH_0064|M.thermoresistible__bu NVEGGNKELERLRKENAEHPIEVTGKKLRQLMSWVDRPITETA
:******:** **: ****.***.**:**:******* ****