For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TARPLSELVEQGWAAALEPVVDQVAEMGRFLRAEIAAGRRYLPAGHSVLRAFTYPFDNVRVLIVGQDPYP TPGHAVGLSFSVAPDVRPLPRSLANVFDEYTADLGYPLPVCGDLTPWAQRGVLLLNRVLTVRPSNPASHR GKGWEVITECAIRALAARSEPMVAILWGRDAATLKPLLTVDNCVVIESPHPSPLSASRGFFGSRPFSRTN EILVGMGAGPINWRLP*
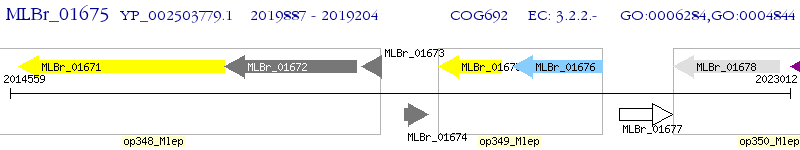
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01675 | ung | - | 100% (227) | uracil-DNA glycosylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3000c | ung | 1e-118 | 88.11% (227) | uracil-DNA glycosylase |
| M. gilvum PYR-GCK | Mflv_4215 | - | 1e-101 | 75.77% (227) | uracil-DNA glycosylase |
| M. tuberculosis H37Rv | Rv2976c | ung | 1e-118 | 88.11% (227) | uracil-DNA glycosylase |
| M. abscessus ATCC 19977 | MAB_3283c | - | 2e-98 | 74.01% (227) | uracil-DNA glycosylase |
| M. marinum M | MMAR_1737 | ung | 1e-111 | 81.94% (227) | uracil-DNA glycosylase Ung |
| M. avium 104 | MAV_3828 | ung | 1e-112 | 83.70% (227) | uracil-DNA glycosylase |
| M. smegmatis MC2 155 | MSMEG_2399 | ung | 1e-106 | 77.97% (227) | uracil-DNA glycosylase |
| M. thermoresistible (build 8) | TH_2052 | ung | 1e-108 | 80.36% (224) | PROBABLE URACIL-DNA GLYCOSYLASE UNG (UDG) |
| M. ulcerans Agy99 | MUL_1980 | ung | 1e-110 | 81.50% (227) | uracil-DNA glycosylase |
| M. vanbaalenii PYR-1 | Mvan_2152 | - | 1e-101 | 75.33% (227) | uracil-DNA glycosylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4215|M.gilvum_PYR-GCK ------------MNARPLSELVDDGWAAALAPVESQVAQMGEFLRAELAA
Mvan_2152|M.vanbaalenii_PYR-1 ------------MTARPLSELIDDGWASALAPVESQVTQMGEFLRAELAD
Mb3000c|M.bovis_AF2122/97 ------------MTARPLSELVERGWAAALEPVADQVAHMGQFLRAEIAA
Rv2976c|M.tuberculosis_H37Rv ------------MTARPLSELVERGWAAALEPVADQVAHMGQFLRAEIAA
MLBr_01675|M.leprae_Br4923 ------------MTARPLSELVEQGWAAALEPVVDQVAEMGRFLRAEIAA
MAV_3828|M.avium_104 ------------MTARPLSELVEPGWARALQPVAEQVARMGQFLRAEIAA
MMAR_1737|M.marinum_M ------------MTARALDELVEPGWAKALEPVADQVTAMGQFLRAELAA
MUL_1980|M.ulcerans_Agy99 ------------MTARALDELVEPGWAKALEPVADQVTAMGQFLRAELAA
MSMEG_2399|M.smegmatis_MC2_155 ------------MTARPLNELVEEGWARALEPVADQVAQMGEFLRTELAE
TH_2052|M.thermoresistible__bu VNERTGVNERTGAAPRPLSELVDDGWARALEPVAGQIREMGDFLRAELAA
MAB_3283c|M.abscessus_ATCC_199 ------------MTARPLAELVDPGWADALGPVADQVTKMGEFLREENSS
.*.* **:: *** ** ** *: ** *** * :
Mflv_4215|M.gilvum_PYR-GCK GHRYLPAGPNILRAFTFPLEKVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
Mvan_2152|M.vanbaalenii_PYR-1 GHRYLPAGENVLRAFTFPLEKVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
Mb3000c|M.bovis_AF2122/97 GRRYLPAGSNVLRAFTFPFDNVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
Rv2976c|M.tuberculosis_H37Rv GRRYLPAGSNVLRAFTFPFDNVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
MLBr_01675|M.leprae_Br4923 GRRYLPAGHSVLRAFTYPFDNVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
MAV_3828|M.avium_104 GRRYLPAGPNVLRAFTYPFDEVKVLIVGQDPYPTPGHAVGLSFSVAPDVS
MMAR_1737|M.marinum_M GRKYLPAGPNVLRAFTFPFDQVRILIVGQDPYPTPGHAVGLSFSVAPDVR
MUL_1980|M.ulcerans_Agy99 GRKYLPAGPNVLRAFTFPFDQVRILIVGQDPYPTPGHAVGLSFSVAPDVR
MSMEG_2399|M.smegmatis_MC2_155 GRQYLPAGQNVLRAFTFPFDQVRVLIVGQDPYPTPGHAVGLSFSVAPDVR
TH_2052|M.thermoresistible__bu GRRYLPAGRNVLRAFTYPFDQVRVLIVGQDPYPTPGHAVGLSFSVAPEVR
MAB_3283c|M.abscessus_ATCC_199 GRGYLPSGANVLRAFTYPLADVRVLIVGQDPYPTPGHAMGLSFSVAADVR
*: ***:* .:*****:*: .*::**************:*******.:*
Mflv_4215|M.gilvum_PYR-GCK PLPRSLDNIFKEYRADLGHPAPSNGDLTPWCEQGVMLLNRVLTVRPGTPA
Mvan_2152|M.vanbaalenii_PYR-1 PLPRSLDNIFREYREDLGYPTPSTGDLTPWCEQGVMLLNRVLTVRPGTPA
Mb3000c|M.bovis_AF2122/97 PWPRSLANIFDEYTADLGYPLPSNGDLTPWAQRGVLLLNRVLTVRPSNPA
Rv2976c|M.tuberculosis_H37Rv PWPRSLANIFDEYTADLGYPLPSNGDLTPWAQRGVLLLNRVLTVRPSNPA
MLBr_01675|M.leprae_Br4923 PLPRSLANVFDEYTADLGYPLPVCGDLTPWAQRGVLLLNRVLTVRPSNPA
MAV_3828|M.avium_104 PLPRSLANIFQEYTADLGHPPPSCGDLTPWAQRGVLLLNRVLTVRPSNPA
MMAR_1737|M.marinum_M PLPRSLSNIFQEYADDLGYPMPACGDLTPWAQRGVMLLNRVLTVRPSNPA
MUL_1980|M.ulcerans_Agy99 PLPRSLSNIFQEYADDLGYPMPACGDLTPWAQRGVMLLNRVLTVRPSNPA
MSMEG_2399|M.smegmatis_MC2_155 PLPRSLSNIFTEYTEDLGHPQPSSGDLTPWAERGVMLLNRVLTVRPGTPA
TH_2052|M.thermoresistible__bu PLPRSLANIFTEYCADLGYPEPSCGDLTPWAERGVLLLNRVLTVQPGSPA
MAB_3283c|M.abscessus_ATCC_199 PVPRSLGNIFDEYQSDLGLPKPANGDLTPWSEQGVMLLNRVLTVRPGNPA
* **** *:* ** *** * * ******.::**:********:*..**
Mflv_4215|M.gilvum_PYR-GCK SHRGKGWEAVTECAIRALVARRQPMVAVLWGRDASTLKPMLE--DTAVIE
Mvan_2152|M.vanbaalenii_PYR-1 SHRGKGWEPVTECAIRALVARQQPMVAVLWGRDASTLKPMLG--DTAVIE
Mb3000c|M.bovis_AF2122/97 SHRGKGWEAVTECAIRALAARAAPLVAILWGRDASTLKPMLAAGNCVAIE
Rv2976c|M.tuberculosis_H37Rv SHRGKGWEAVTECAIRALAARAAPLVAILWGRDASTLKPMLAAGNCVAIE
MLBr_01675|M.leprae_Br4923 SHRGKGWEVITECAIRALAARSEPMVAILWGRDAATLKPLLTVDNCVVIE
MAV_3828|M.avium_104 SHRGKGWEPVTECAIRALTARQQPLVAILWGRDASTLKPILAQGNCVAIE
MMAR_1737|M.marinum_M SHRGKGWEAVTECAIRALAARSQPMVAILWGRDASTLKPMLADGDCASIE
MUL_1980|M.ulcerans_Agy99 SHRGKGWEAVTECAIRALAARSQPMVAILWGRDASTLKSMLAGGDCASIE
MSMEG_2399|M.smegmatis_MC2_155 SHRGKGWEAVTECAIRALVAREQPLVAVLWGRDAATLKPMLAEGDCATIE
TH_2052|M.thermoresistible__bu SHRGKGWEAVTECAIRALVARRRPLVAVLWGRDAATLRPMLEGENCVAIE
MAB_3283c|M.abscessus_ATCC_199 SHRGKGWEIVTECAIRALVARDAPLVAILWGRDAATLKPMLG-PSVPTIE
******** :********.** *:**:******:**:.:* . **
Mflv_4215|M.gilvum_PYR-GCK SPHPSPLSASRGFFGSKPFSRANELLAQRGAEPVDWRLP
Mvan_2152|M.vanbaalenii_PYR-1 SPHPSPLSASRGFFGSKPFSRANELLTQMGAEPVDWRLP
Mb3000c|M.bovis_AF2122/97 SPHPSPLSASRGFFGSRPFSRANELLVGMGAEPIDWRLP
Rv2976c|M.tuberculosis_H37Rv SPHPSPLSASRGFFGSRPFSRANELLVGMGAEPIDWRLP
MLBr_01675|M.leprae_Br4923 SPHPSPLSASRGFFGSRPFSRTNEILVGMGAGPINWRLP
MAV_3828|M.avium_104 SPHPSPLSASRGFFGSRPFSRANKLLAEMGADEIDWRLP
MMAR_1737|M.marinum_M SPHPSPLSASRGFFGSRPFSRANELLTGRGADPIDWRLP
MUL_1980|M.ulcerans_Agy99 SPHPSPLSASRGFFGSRPFSRANELLTGRGADPIDWRLP
MSMEG_2399|M.smegmatis_MC2_155 SPHPSPLSASRGFFGSRPFSRVNELLQRRGIEPIDWKLP
TH_2052|M.thermoresistible__bu SPHPSPLSASRGFFGSRPFSRANELLTRLGGEPVDWRLP
MAB_3283c|M.abscessus_ATCC_199 SVHPSPLSASRGFFGSKPFSRANELLVGLGAQPVDWRLP
* **************:****.*::* * ::*:**