For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVALALRDGCRRISTMTRQYGVTTQRHLISPVVFDITPLGRRPGAIIALQKTVPSLARIGLELVVIEWGA PINLDLRVESVSEDVLVAGTVTAPTVSECVRCLTAVHGHVQVTLNQLFAYPYSATKVTTEEDAVGHVVDG TIDLEQSIIDAVGIELPFAPMCRSDCPGLCAECGTSLVVEPGHPHDRIDPWWAKLTDMLAPDVPQTSETD GSRSEW*
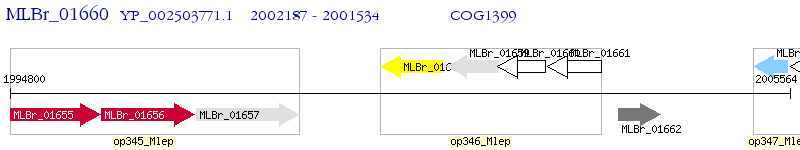
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01660 | - | - | 100% (217) | hypothetical protein MLBr_01660 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2951c | - | 6e-71 | 67.19% (192) | hypothetical protein Mb2951c |
| M. gilvum PYR-GCK | Mflv_4193 | - | 5e-53 | 54.55% (187) | hypothetical protein Mflv_4193 |
| M. tuberculosis H37Rv | Rv2926c | - | 6e-71 | 67.19% (192) | hypothetical protein Rv2926c |
| M. abscessus ATCC 19977 | MAB_3257c | - | 7e-49 | 53.53% (170) | hypothetical protein MAB_3257c |
| M. marinum M | MMAR_1781 | - | 1e-62 | 59.90% (192) | hypothetical protein MMAR_1781 |
| M. avium 104 | MAV_3784 | - | 5e-68 | 69.01% (171) | hypothetical protein MAV_3784 |
| M. smegmatis MC2 155 | MSMEG_2417 | - | 1e-53 | 51.24% (201) | hypothetical protein MSMEG_2417 |
| M. thermoresistible (build 8) | TH_1225 | - | 6e-49 | 52.97% (185) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2029 | - | 8e-63 | 59.90% (192) | hypothetical protein MUL_2029 |
| M. vanbaalenii PYR-1 | Mvan_2170 | - | 2e-52 | 52.66% (188) | hypothetical protein Mvan_2170 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01660|M.leprae_Br4923 MTVALALRDGCRRISTMTRQYGVTTQRHLISPVVFDITPL--GRRPGAII
MAV_3784|M.avium_104 --------------------------------MTFDITRL--GRRPGAMV
Mb2951c|M.bovis_AF2122/97 ----MDLGGVRRRISLMARQHGPTAQRHVASPMTVDIARL--GRRPGAMF
Rv2926c|M.tuberculosis_H37Rv ----MDLGGVRRRISLMARQHGPTAQRHVASPMTVDIARL--GRRPGAMF
MMAR_1781|M.marinum_M ----------------MARQRSQTKSGHVSSPLTVDIARL--GRRPGSMV
MUL_2029|M.ulcerans_Agy99 ----------------MARQRSQTKSGHVSSPLTVDIARL--GRRPGSMV
MAB_3257c|M.abscessus_ATCC_199 ----------------------------MPGPFVLDVLALGLGRRPGSMR
Mflv_4193|M.gilvum_PYR-GCK ----------------MATHASAAARRSSRSPLVIDITRL--GRRPGSMI
Mvan_2170|M.vanbaalenii_PYR-1 ----------------MARYVNAAAHRGSRSPLVIDISRL--GRRPGSMM
MSMEG_2417|M.smegmatis_MC2_155 ----------------MATHANAAKHRESRSPLVLDVSRL--GRRPGSML
TH_1225|M.thermoresistible__bu ---------------------------------VINISRL--GRQPGSML
..:: * **:**::
MLBr_01660|M.leprae_Br4923 ALQKTVPSLARIGLELVVIEWGAPINLDLRVESVSEDVLVAGTVTAPTVS
MAV_3784|M.avium_104 TLRTTVPSPARIGLDMIAIEAGAPLDLDLQVQSVSEGVLVTGSVTGPTVG
Mb2951c|M.bovis_AF2122/97 ELHDTVHSPARIGLELIAIDQGALLDLDLRVESVSEGVLVTGTVAAPTVG
Rv2926c|M.tuberculosis_H37Rv ELHDTVHSPARIGLELIAIDQGALLDLDLRVESVSEGVLVTGTVAAPTVG
MMAR_1781|M.marinum_M TVHTTVDSPSRIGLDLIAIDKGAPVELDLRVESVSEGVLVTGTVAADTVG
MUL_2029|M.ulcerans_Agy99 TVHTTVDSPSRIGLDLIAIDKGAPVELDLRVESVSEGVLVTGTVAADTVG
MAB_3257c|M.abscessus_ATCC_199 EIERTFASPVRIGNDLIAIPEGTDVDMDLRVESVSEGVLVTGTITGDLAG
Mflv_4193|M.gilvum_PYR-GCK TVETTVPSPTRIGVELIAIEEGAPLELDLRLESVSEGVLVTGTMSAPTSG
Mvan_2170|M.vanbaalenii_PYR-1 TVEETVPSPVRMGVELIAVAEGAPLELNLRLESVSEGVLVTGTVWAPTTG
MSMEG_2417|M.smegmatis_MC2_155 THRETVPSPARIGLDLIAIRPGAELDLDLRLESVSEGVLVTGTVSAHTEG
TH_1225|M.thermoresistible__bu TFDETVASPSRLGVALVGVEQGAPLEFHLRIEAVSEGVLVSGTMSAPTTG
*. * *:* :: : *: :::.*::::***.***:*:: . .
MLBr_01660|M.leprae_Br4923 ECVRCLTAVHGHVQVTLNQLFAYPYSATKVTTEEDAVGHVVD----GTID
MAV_3784|M.avium_104 ECARCLTALDGEVRVRLTELFAYPDSTTEATTEEDEVGHIVD----DTVD
Mb2951c|M.bovis_AF2122/97 ECARCLSPVRGRVQVALTELFAYPDSATDETTEEDEVGRVVD----ETID
Rv2926c|M.tuberculosis_H37Rv ECARCLSPVRGRVQVALTELFAYPDSATDETTEEDEVGRVVD----ETID
MMAR_1781|M.marinum_M ECSRCLTPVTGSVQVHLTELFAYPDSTTEATTEEGEVSRVVD----ERID
MUL_2029|M.ulcerans_Agy99 ECSRCLTPVTGSVQVHLTELFAYPDSTTEATTEEGEVSRVVD----ERID
MAB_3257c|M.abscessus_ATCC_199 ECSRCLEPITGHADATITELFAYPDSETEATTEEDEVHRVVD----GLVD
Mflv_4193|M.gilvum_PYR-GCK ECGRCLKPITGEVSIDLTELFAYPDSTTDETTESDELPRVGRERGTDTVD
Mvan_2170|M.vanbaalenii_PYR-1 ECARCLTPITGEVEIDLTELFAYPDSTTEETTEADEIPRVSRDRGGEGVD
MSMEG_2417|M.smegmatis_MC2_155 ECARCLTAISGDVEIDLTELYAYPDSTTDETSESDEIARVGASGRPDTVD
TH_1225|M.thermoresistible__bu ECARCLDPVHGRVDIELTELFAYPDSITAETTDADDLGRVDTTGDVDTVD
** *** .: * . :.:*:*** * * *:: . : :: :*
MLBr_01660|M.leprae_Br4923 LEQSIIDAVGIELPFAPMCRSDCPGLCAECGTSLV-VEPGHPHDRIDPWW
MAV_3784|M.avium_104 LEQSIIDAVGLELPFSPVCRPDCPGLCPECGVSLA-AEPGHQHDRIDPRW
Mb2951c|M.bovis_AF2122/97 LEQPIIDAVGLELPFSPVCRPDCPGLCPQCGVPLA-SEPGHRHEQIDPRW
Rv2926c|M.tuberculosis_H37Rv LEQPIIDAVGLELPFSPVCRPDCPGLCPQCGVPLA-SEPGHRHEQIDPRW
MMAR_1781|M.marinum_M LEQPIIDAVGLELPLSPVCRPDCSGLCPQCGVALD-AEPGHHHDQIDPRW
MUL_2029|M.ulcerans_Agy99 LEQPIIDAVGLELPLSPVCRPDCSGLCPQCGVALD-AEPGHHHDQIDPRW
MAB_3257c|M.abscessus_ATCC_199 LEQTIVDAIVPDLELAPVCRPDCPGLCPDCGIALATAEPGHHHDKIDPRW
Mflv_4193|M.gilvum_PYR-GCK LEQSVIDAVGLALPFSPVCGPDCQGLCPDCGVALEDAEPGHHHEKIDPRW
Mvan_2170|M.vanbaalenii_PYR-1 LEQSIVDAVGLVLPFSPVCGPDCQGLCPECGVRLADAEPGHHHEQIDPRW
MSMEG_2417|M.smegmatis_MC2_155 LEQPIVDAVGMSLPFAPLCQPDCPGLCPDCGIPLATAEPGHGHEKIDPRW
TH_1225|M.thermoresistible__bu LEQPIIDAVGLVLPFSPLCGPDCLGLCPQCGIRLATAEPGHRHEVIDPRW
***.::**: * ::*:* .** ***.:** * **** *: *** *
MLBr_01660|M.leprae_Br4923 AKLTDMLAPDVPQTSETDGSRSEW-
MAV_3784|M.avium_104 AKLAGMFA--------SDEARDER-
Mb2951c|M.bovis_AF2122/97 AKLVEMLGP------ESDTLRGER-
Rv2926c|M.tuberculosis_H37Rv AKLVEMLGP------ESDTLRGER-
MMAR_1781|M.marinum_M SKLADMFTPD----GPEATGRGER-
MUL_2029|M.ulcerans_Agy99 SKLADMFTPD----GPEATGRGER-
MAB_3257c|M.abscessus_ATCC_199 AKLANFGD---------DQ------
Mflv_4193|M.gilvum_PYR-GCK AKLAEFGEK-------DGDDA----
Mvan_2170|M.vanbaalenii_PYR-1 AKLAQLREQS------AGDDS----
MSMEG_2417|M.smegmatis_MC2_155 AKLAGMLQDDSKDRDRAGGDA----
TH_1225|M.thermoresistible__bu AKLAAMFPDREAVSDRDGDGPGESG
:**. :