For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSLIDHLTELRTRLLISLAAIVVTTIFGFIWYSHSIFGLESLGEWLRRPYCSLPQSARADISPDGQCRLL ATAPFDQFMLRIKVGMAAGIVLASPVWFYQLWAFITPGLYTKERRFTVAFVVPAAVLFAGGTVLAYLVLS KALGFLLIVGSGVQVTALSGDRYFGFLLNLLVVFGVSFEFPLLIVMLNIAGLLTYQRLKSWRRGLIFAMF VFAAVFTPGSDPFSMTALGAALTVLLELAIQLVRLHDKRRVKHEALIADDEASVIEPPSSIPERSYTATR SHDDVT
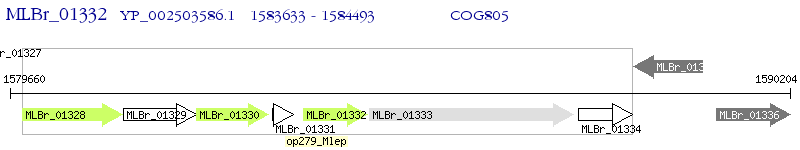
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01332 | - | - | 100% (286) | membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2120c | tatC | 1e-140 | 86.01% (286) | Sec-independent protein translocase transmembrane protein |
| M. gilvum PYR-GCK | Mflv_3094 | - | 1e-110 | 68.50% (273) | Sec-independent protein translocase, TatC subunit |
| M. tuberculosis H37Rv | Rv2093c | tatC | 1e-140 | 86.01% (286) | Sec-independent protein translocase transmembrane protein |
| M. abscessus ATCC 19977 | MAB_2188 | - | 1e-112 | 69.60% (273) | Sec-independent protein translocase protein TatC-like protein |
| M. marinum M | MMAR_3079 | tatC | 1e-137 | 84.27% (286) | sec-independent protein translocase transmembrane protein |
| M. avium 104 | MAV_2410 | tatC | 1e-133 | 80.90% (288) | twin arginine-targeting protein translocase TatC |
| M. smegmatis MC2 155 | MSMEG_3886 | tatC | 1e-113 | 68.26% (293) | twin arginine-targeting protein translocase TatC |
| M. thermoresistible (build 8) | TH_0502 | - | 1e-111 | 67.69% (294) | Probable Sec-independent protein translocase transmembrane |
| M. ulcerans Agy99 | MUL_2325 | tatC | 1e-136 | 84.27% (286) | sec-independent protein translocase transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_3441 | - | 1e-113 | 69.96% (283) | Sec-independent protein translocase, TatC subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3094|M.gilvum_PYR-GCK ----------MPGLFKKLDPRRRRSRVNPDGTMSLVDHLTELRSRLLIAV
Mvan_3441|M.vanbaalenii_PYR-1 --------MQIPGILKKLDPRQRRSRVNPDGTMSLVDHLTELRSRLLIAV
MSMEG_3886|M.smegmatis_MC2_155 --------MQTPGIFKKLDPRRRRSRVNPDGTMSLVDHLHELRNRLLISV
TH_0502|M.thermoresistible__bu --------VRTSGLFTKLDPRRRRARVNPDGTMSLVEHLQELRNRLLISL
MMAR_3079|M.marinum_M MVNGFRASARAGALFKRLNPRMRRSRVNPDGTMSLVDHLTELRTRLLISL
MUL_2325|M.ulcerans_Agy99 -MNGFRASARAGALFKRLNPRMRRSRVNPDGTMSLVDHLTELRTRLLISL
MLBr_01332|M.leprae_Br4923 --------------------------------MSLIDHLTELRTRLLISL
Mb2120c|M.bovis_AF2122/97 --------MRAAGLLKRLNPRNRRSRVNPDATMSLVDHLTELRTRLLISL
Rv2093c|M.tuberculosis_H37Rv --------MRAAGLLKRLNPRNRRSRVNPDATMSLVDHLTELRTRLLISL
MAV_2410|M.avium_104 -MKAFKVSARTSGLLKRLNPRNRRSRTNPDATMSLVDHLTELRTRLLISL
MAB_2188|M.abscessus_ATCC_1997 -------MNKVLGALR--LPGRPRQPKNPDGTMSLVEHLQELRTRLLISV
***::** ***.****::
Mflv_3094|M.gilvum_PYR-GCK LAVLATSVLGFLWYSNGFLGLPSLGDWLRGPYCALPDSARASITPDGECR
Mvan_3441|M.vanbaalenii_PYR-1 LAVLVTTILGFLWYSHGVLGLPSLGDWLRGPYCALPDSARASITPDGGCR
MSMEG_3886|M.smegmatis_MC2_155 AAVVLTTIIGFIWYTHGVFGFNSLGEWLRGPYCSLPDSARANIAPDGECR
TH_0502|M.thermoresistible__bu AAVVLATVVGFVWYSHGVFTLPSLGEWLRGPYCALPESARADITTDGGCR
MMAR_3079|M.marinum_M AAILLTTIFGYIWYSHPVFGLESLGEWLRQPYCSLPQSARADISADGQCR
MUL_2325|M.ulcerans_Agy99 AAILLTTIFGYIWYSHPVFGLESLGEWLRQPYCSLPQSARADISADGQCR
MLBr_01332|M.leprae_Br4923 AAIVVTTIFGFIWYSHSIFGLESLGEWLRRPYCSLPQSARADISPDGQCR
Mb2120c|M.bovis_AF2122/97 AAILVTTIFGFVWYSHSIFGLDSLGEWLRHPYCALPQSARADISADGECR
Rv2093c|M.tuberculosis_H37Rv AAILVTTIFGFVWYSHSIFGLDSLGEWLRHPYCALPQSARADISADGECR
MAV_2410|M.avium_104 AAIALTTIFGFFWYSHSIFGLESLGEWLRHPYCALPQSARAEISANGQCR
MAB_2188|M.abscessus_ATCC_1997 AAVLLTTIVGFVWYEHTLFGIESLGEWLREPYCSLPASARANLSVDGACR
*: :::.*:.** : .: : ***:*** ***:** ****.:: :* **
Mflv_3094|M.gilvum_PYR-GCK LLATAPFDQFMLRLKVAMTAGVVMACPVWLYQVWAFITPGLYAKERRFAV
Mvan_3441|M.vanbaalenii_PYR-1 LLATAPFDQFMLRLKVGMTAGIVLACPVWLYQLWAFITPGLYAKERRFAV
MSMEG_3886|M.smegmatis_MC2_155 LLATAPFDQFMLRLKVALAAGVVLACPVWLYQLWAFITPGLYKKERRFAM
TH_0502|M.thermoresistible__bu LLATSPFDQFLLRLKVALAAGLVLACPVWLYQLWAFITPALYKNERRFAV
MMAR_3079|M.marinum_M LLATAPFDQFMLRLKVGMAAGIVLACPIWFYELWAFITPGLYQKERRFAV
MUL_2325|M.ulcerans_Agy99 LLATAPFDQFMLRLKVGMAAGIVLACPIWFYELWAFITPGLYQKERRFAV
MLBr_01332|M.leprae_Br4923 LLATAPFDQFMLRIKVGMAAGIVLASPVWFYQLWAFITPGLYTKERRFTV
Mb2120c|M.bovis_AF2122/97 LLATAPFDQFMLRLKVGMAAGIVLACPVWFYQLWAFITPGLYQRERRFAV
Rv2093c|M.tuberculosis_H37Rv LLATAPFDQFMLRLKVGMAAGIVLACPVWFYQLWAFITPGLYQRERRFAV
MAV_2410|M.avium_104 LLATAPFDQFMLRLKVGLTAGVVLACPVWFYQLWAFITPGLYQKERRFAI
MAB_2188|M.abscessus_ATCC_1997 LLATAPFEQFMLRLKVGLTAGVVLACPVWLYQIWRFITPGLYRNERKFAI
****:**:**:**:**.::**:*:*.*:*:*::* ****.** .**:*::
Mflv_3094|M.gilvum_PYR-GCK AFVMVGAILFVAGAILAYVVLSTALGFLLTVGSDVQVTALSGDQYFGFLI
Mvan_3441|M.vanbaalenii_PYR-1 AFVMVGAVLFVAGAILAYVVLSKALGFLLTVGSDVQVTALSGDQYFGFLI
MSMEG_3886|M.smegmatis_MC2_155 AFVSFGAMLFISGAVLAYVVLSKALSFLLTVGSDVQVTALSGDQYFGFLI
TH_0502|M.thermoresistible__bu AFVTVGAALFIAGAVLAYLVLATALGFLLTVGSDVQVTALSGDQYFGFLV
MMAR_3079|M.marinum_M AFVIPAALLFVAGAVLAYLVLSKALSFLLTVGSDVQVTALSGDRYFGFLI
MUL_2325|M.ulcerans_Agy99 AFVIPAALLFVAGAVLAYLVLSKALSFLLTVGSDVQVTALSGDRYFGFLI
MLBr_01332|M.leprae_Br4923 AFVVPAAVLFAGGTVLAYLVLSKALGFLLIVGSGVQVTALSGDRYFGFLL
Mb2120c|M.bovis_AF2122/97 AFVIPAAVLFVAGAVLAYLVLSKALGFLLTVGSDVQVTALSGDRYFGFLL
Rv2093c|M.tuberculosis_H37Rv AFVIPAAVLFVAGAVLAYLVLSKALGFLLTVGSDVQVTALSGDRYFGFLL
MAV_2410|M.avium_104 AFVIPAVALFVAGAVLAYFVLSKALGFLLTVGSDVQVTALSGDRYFGFLI
MAB_2188|M.abscessus_ATCC_1997 IFVTAAAVLFVAGALLAYLVLAKALHFLLTVGSDVQVTGLKGDEYFSFLI
** .. ** .*::***.**:.** *** ***.****.*.**.**.**:
Mflv_3094|M.gilvum_PYR-GCK NLLLVFGISFEFPLLIIMLNLVGILTYERLKAWRRGLIMGLFVFCAFVTP
Mvan_3441|M.vanbaalenii_PYR-1 NLLLVFGISFEFPLLIIMLNLVGILTYERLKAWRRGLIMGVFVFCAFATP
MSMEG_3886|M.smegmatis_MC2_155 NLLLVFGISFEFPLLIIMLNLVGILTYERLSAWRRGLIFGLFVFAAFATP
TH_0502|M.thermoresistible__bu NLLLVFGFSFEFPLLIIMLNLVGVLTYERLKAWRRGLIFTMFVFAAIFTP
MMAR_3079|M.marinum_M NLLVVFGVSFEFPLLIVMLNMAGLLTYQRLKSWRRGLIFAMFVFAAVFTP
MUL_2325|M.ulcerans_Agy99 NLLVVFGVSFEFPLLIVMLNMAGLLTYQRLKSWRRGLIFAMFVFAAVFTP
MLBr_01332|M.leprae_Br4923 NLLVVFGVSFEFPLLIVMLNIAGLLTYQRLKSWRRGLIFAMFVFAAVFTP
Mb2120c|M.bovis_AF2122/97 NLLVVFGVSFEFPLLIVMLNLAGLLTYERLKSWRRGLIFAMFVFAAIFTP
Rv2093c|M.tuberculosis_H37Rv NLLVVFGVSFEFPLLIVMLNLAGLLTYERLKSWRRGLIFAMFVFAAIFTP
MAV_2410|M.avium_104 NLLIVFGVSFEFPLLIVMLNTIGVLTYPRLKAWRRGLIFAMFVFAAIFTP
MAB_2188|M.abscessus_ATCC_1997 NLLLVFGVSFEFPLLIVMLNFVGILSYDKLRNWRRGLIFGLFVFAAFATP
***:***.********:*** *:*:* :* ******: :***.*. **
Mflv_3094|M.gilvum_PYR-GCK GSDPFSMLALALAMTALMEVAIQVARISDKRKARRAALDDVPDDEASPIG
Mvan_3441|M.vanbaalenii_PYR-1 GSDPFSMLALALAMTLLMELAIQVARLSDRRKARRAALDAVDDDEAAPIG
MSMEG_3886|M.smegmatis_MC2_155 GSDPFSMLALACALTVLLEFAIQIARLNDRRKARRAAME-VPEDEAADIG
TH_0502|M.thermoresistible__bu GSDPFSMLALALALTLLLEFAIQIARFNDRRRAARARLEEVPDEQAAPIG
MMAR_3079|M.marinum_M GSDPFSMTALGVALTVLLEFAIQIARLHDKRKAKREV--LLADDEAS---
MUL_2325|M.ulcerans_Agy99 GSDPFSMTALGVALTVLLEFAIQIARLHDKRKAKREV--LPADDEAS---
MLBr_01332|M.leprae_Br4923 GSDPFSMTALGAALTVLLELAIQLVRLHDKRRVKHEA--LIADDEAS---
Mb2120c|M.bovis_AF2122/97 GSDPFSMTALGAALTVLLELAIQIARVHDKRKAKREA--AIPDDEAS---
Rv2093c|M.tuberculosis_H37Rv GSDPFSMTALGAALTVLLELAIQIARVHDKRKAKREA--AIPDDEAS---
MAV_2410|M.avium_104 GSDPFSMTALGVALSVLLEFAIQIARLHDKRKAKREAQAVIPDDQAS---
MAB_2188|M.abscessus_ATCC_1997 GSDPFSMTALGLALTLLLEFSIQVSRVHDKRKAKREAAQQLSDDEAS---
******* **. *:: *:*.:**: *. *:*:. : :::*:
Mflv_3094|M.gilvum_PYR-GCK PAESIDAPTAVP--APSRVVVDDEST--
Mvan_3441|M.vanbaalenii_PYR-1 PVETIEAPTAVP--APSRLVVDDDAT--
MSMEG_3886|M.smegmatis_MC2_155 SVEPIEPPAPVSGTTAGRATVDDDAT--
TH_0502|M.thermoresistible__bu EVEPIEPPEPVR--GPLRDAIHDDVT--
MMAR_3079|M.marinum_M ---SIEEPEPIPTPSAPGG-SHDDIT--
MUL_2325|M.ulcerans_Agy99 ---SIEEPEPIPTPSAPGG-SHDDIT--
MLBr_01332|M.leprae_Br4923 ---VIEPPSSIPERSYTATRSHDDVT--
Mb2120c|M.bovis_AF2122/97 ---VIDPPSPVPAPSV-IG-SHDDVT--
Rv2093c|M.tuberculosis_H37Rv ---VIDPPSPVPAPSV-IG-SHDDVT--
MAV_2410|M.avium_104 ---VIEPPTAIPEPSLSAG-QHDDVT--
MAB_2188|M.abscessus_ATCC_1997 ---TLDAPEAIDAPEPIGPAERFDNRGK
:: * .: :