For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPTDYDAPRRTETDNVPEDSLEELKARRNEAASAVVDVDESESAESFELPGADLSGEELSVRVIPKQADE FTCSSCFLVQHRSRLASEKNGVMICTDCTA
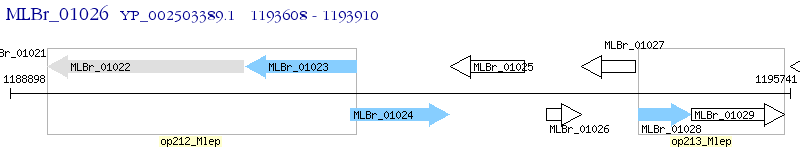
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01026 | - | - | 100% (100) | hypothetical protein MLBr_01026 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2718c | - | 1e-51 | 96.00% (100) | hypothetical protein Mb2718c |
| M. gilvum PYR-GCK | Mflv_3935 | - | 3e-49 | 93.00% (100) | hypothetical protein Mflv_3935 |
| M. tuberculosis H37Rv | Rv2699c | - | 1e-51 | 96.00% (100) | hypothetical protein Rv2699c |
| M. abscessus ATCC 19977 | MAB_3005c | - | 2e-46 | 87.00% (100) | hypothetical protein MAB_3005c |
| M. marinum M | MMAR_2015 | - | 2e-50 | 94.00% (100) | hypothetical protein MMAR_2015 |
| M. avium 104 | MAV_3591 | - | 3e-51 | 96.00% (100) | hypothetical protein MAV_3591 |
| M. smegmatis MC2 155 | MSMEG_2763 | - | 1e-48 | 91.00% (100) | hypothetical protein MSMEG_2763 |
| M. thermoresistible (build 8) | TH_1190 | - | 6e-48 | 89.00% (100) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3337 | - | 1e-50 | 94.00% (100) | hypothetical protein MUL_3337 |
| M. vanbaalenii PYR-1 | Mvan_2465 | - | 4e-49 | 93.00% (100) | hypothetical protein Mvan_2465 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3935|M.gilvum_PYR-GCK --MATDYDAPRRTETDDVSEDSLEELKARRNEAQSAVVDVDESESAESFE
Mvan_2465|M.vanbaalenii_PYR-1 --MATDYDAPRRTETDDVSEDSLEELKARRNEAQSAVVDVDESESAENFE
Mb2718c|M.bovis_AF2122/97 --MPTDYDAPRRTETDDVSEDSLEELKARRNEAASAVVDVDESESAESFE
Rv2699c|M.tuberculosis_H37Rv --MPTDYDAPRRTETDDVSEDSLEELKARRNEAASAVVDVDESESAESFE
MLBr_01026|M.leprae_Br4923 --MPTDYDAPRRTETDNVPEDSLEELKARRNEAASAVVDVDESESAESFE
MMAR_2015|M.marinum_M --MPTDYDAPRRTESDDVSEDSLEELKARRNEAASSVVDVDESESAESFE
MUL_3337|M.ulcerans_Agy99 --MPTDYDAPRRTESDDVSEDSLEELKARRNEAASSVVDVDESESAESFE
MAV_3591|M.avium_104 --MPTDYDAPRRTETDDVSEDSLEELKARRNEAASSVVDVDESESAESFE
TH_1190|M.thermoresistible__bu MAMATDYDAPRRTETDDVSEDSLEELKARRNEAQSAVVDVDDSETVESYE
MSMEG_2763|M.smegmatis_MC2_155 --MATDYDAPRRTETDDVSEDSLEELKARRNEAQSAVVDVDEAETAESFE
MAB_3005c|M.abscessus_ATCC_199 --MATDYDAPRRSDADDVSEDSLEELKARRNEAQSAVVDVDEAETAESFE
*.********:::*:*.************** *:*****::*:.*.:*
Mflv_3935|M.gilvum_PYR-GCK LPGADLSGEELSVRVIPKQADEFTCSSCFLVHHRSRLASEKNGMMICTDC
Mvan_2465|M.vanbaalenii_PYR-1 LPGADLSGEELSVRVIPKQADEFTCSSCFLVHHRSRLASEKNGVMICTDC
Mb2718c|M.bovis_AF2122/97 LPGADLSGEELSVRVVPKQADEFTCSSCFLVQHRSRLASEKNGVMICTDC
Rv2699c|M.tuberculosis_H37Rv LPGADLSGEELSVRVVPKQADEFTCSSCFLVQHRSRLASEKNGVMICTDC
MLBr_01026|M.leprae_Br4923 LPGADLSGEELSVRVIPKQADEFTCSSCFLVQHRSRLASEKNGVMICTDC
MMAR_2015|M.marinum_M LPGADLSGEELSVRVIPKQADEFTCSSCFLVQHRSRLASEKNGLMICTDC
MUL_3337|M.ulcerans_Agy99 LPGADLSGEELSVRVIPKQADEFTCSSCFLVQHRSRLASEKNGLMICTDC
MAV_3591|M.avium_104 LPGADLSGEELSVRVIPKQADEFTCSSCFLVQHRSRLASEKNGVMICTDC
TH_1190|M.thermoresistible__bu LPGADLSGEELSVRVIPKQADEFTCSSCFLVHHRSRLASEKNGVMLCTDC
MSMEG_2763|M.smegmatis_MC2_155 LPGADLSGEELSVRVVPKQADEFTCSSCFLVHHRSRLASEKNGVMICTDC
MAB_3005c|M.abscessus_ATCC_199 LPGADLSGEELSVRVIPKQADEFTCSSCFLVHHRSRLASEKNGQLICTDC
***************:***************:*********** ::****
Mflv_3935|M.gilvum_PYR-GCK AA
Mvan_2465|M.vanbaalenii_PYR-1 AA
Mb2718c|M.bovis_AF2122/97 AA
Rv2699c|M.tuberculosis_H37Rv AA
MLBr_01026|M.leprae_Br4923 TA
MMAR_2015|M.marinum_M AA
MUL_3337|M.ulcerans_Agy99 AA
MAV_3591|M.avium_104 AA
TH_1190|M.thermoresistible__bu AA
MSMEG_2763|M.smegmatis_MC2_155 AA
MAB_3005c|M.abscessus_ATCC_199 AA
:*