For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTLNTIALELVPPNLDAGQERAVEDACKVVQYSAESGLSGRIRHVMIPGMIVEEDDRPIPMKPKLDVLDF WSIVRPELPGVNGLCTQVTAFMDEASLGRRLADLSAAGMEGVVFVGVPRTMNDGEGSGVAPTDALSMYRE LVANRGVIVIPTRDGEQDRLNFKCNQGATYGMTQLLYSDAIVRFLTEFAKHTDHRPELLLSFGFVPKAER RVGLINWLIQDSGNAAVADEQEFVKRLAGSEPAQKRQLMLDLYQRVIDGVAELGFPLSIHFEATYGMSPA AFDTFAEMVAYWPPVCVGQSGEST
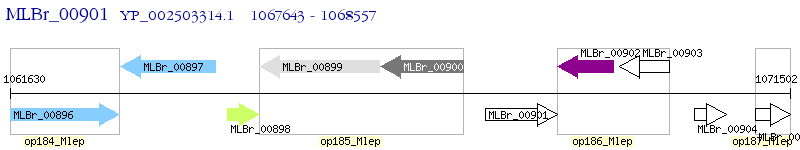
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00901 | - | - | 100% (304) | hypothetical protein MLBr_00901 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2194c | - | 1e-142 | 81.27% (299) | hypothetical protein Mb2194c |
| M. gilvum PYR-GCK | Mflv_1061 | - | 1e-128 | 74.49% (294) | hypothetical protein Mflv_1061 |
| M. tuberculosis H37Rv | Rv2172c | - | 1e-142 | 81.27% (299) | hypothetical protein Rv2172c |
| M. abscessus ATCC 19977 | MAB_4622c | - | 1e-114 | 65.41% (292) | hypothetical protein MAB_4622c |
| M. marinum M | MMAR_3207 | - | 1e-143 | 82.99% (294) | hypothetical protein MMAR_3207 |
| M. avium 104 | MAV_2322 | - | 1e-138 | 80.61% (294) | hypothetical protein MAV_2322 |
| M. smegmatis MC2 155 | MSMEG_6596 | - | 1e-123 | 71.09% (294) | hypothetical protein MSMEG_6596 |
| M. thermoresistible (build 8) | TH_1200 | - | 1e-126 | 72.79% (294) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3515 | - | 1e-143 | 82.65% (294) | hypothetical protein MUL_3515 |
| M. vanbaalenii PYR-1 | Mvan_5770 | - | 1e-127 | 73.13% (294) | hypothetical protein Mvan_5770 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1061|M.gilvum_PYR-GCK MWSAAVSLNTIALELVPPNVERGREQALEDAEKVLRCSAEAGLAGRIRHV
Mvan_5770|M.vanbaalenii_PYR-1 -----MSLNTIALELVPPNVERGREQAMEDAEKVLRCSAEAGLAGRIRHV
MSMEG_6596|M.smegmatis_MC2_155 MGSRFVPLNTIALELVPPNIERGASYAVEEAHKVLAIAAETGIEGRIRHV
MMAR_3207|M.marinum_M -----MTLRTIALELVPPNVDGGKERALEDARKVVQYSADFGLDGRLRHV
MUL_3515|M.ulcerans_Agy99 -----MTLRTIALELVPPNVDGGKERALEDARKVVQYSADFGLDGRLRHV
MLBr_00901|M.leprae_Br4923 -----MTLNTIALELVPPNLDAGQERAVEDACKVVQYSAESGLSGRIRHV
Mb2194c|M.bovis_AF2122/97 -----MTLNTIALELVPPNLEGGKERAIEDARKVVQYSAASGLDGRIRHV
Rv2172c|M.tuberculosis_H37Rv -----MTLNTIALELVPPNLEGGKERAIEDARKVVQYSAASGLDGRIRHV
MAV_2322|M.avium_104 -----MTLNTIALELVPPNADEGRERALEEAHKVVRHSAESGLDGRIRHV
TH_1200|M.thermoresistible__bu -----VTLNTVALELVPPNVERGAEQALEEAHKVLRLSAQSGLDGRIKHV
MAB_4622c|M.abscessus_ATCC_199 -------MNTVALEMVPPNVDGGLDRAAEELRKLRQFSAEFGIDRRIAHV
:.*:***:**** : * . * *: *: :* *: *: **
Mflv_1061|M.gilvum_PYR-GCK MIPGMIEEDGDRPIEMKPKLDVLDFWSMIKPELPDVKGLCTQVTAFMDEE
Mvan_5770|M.vanbaalenii_PYR-1 MIPGMIEEDGDRPIEMKPKLDVLDFWNMIRPELPGIKGLCTQVTAFMDEA
MSMEG_6596|M.smegmatis_MC2_155 MIPGMIDEDDDRPIEMKPKMDVLEYWNIIRPELPGLRGLCTQVTSFLGEE
MMAR_3207|M.marinum_M MIPGMIAEDDDRPVPMQQKLDVLDFWQIIKPEMPGINGLCTQVTAFTDEA
MUL_3515|M.ulcerans_Agy99 MIPGMIAEDDDRPVPMQQKLDVLDFWQIIKPEMPGINGLCTQVTAFTDEA
MLBr_00901|M.leprae_Br4923 MIPGMIVEEDDRPIPMKPKLDVLDFWSIVRPELPGVNGLCTQVTAFMDEA
Mb2194c|M.bovis_AF2122/97 MMPGMIAEDDDRPIPMQPKLDVLDFWSIIKPELAGVHGLCTQVTAFMDEP
Rv2172c|M.tuberculosis_H37Rv MMPGMIAEDDDRPIPMQPKLDVLDFWSIIKPELAGVHGLCTQVTAFMDEP
MAV_2322|M.avium_104 MIPGIIAEDDDRPVPMKPKLDVLDFWSVVKPELPGIKGLCTQVTAFMDEQ
TH_1200|M.thermoresistible__bu MIPGMIEEDDDRPVPMKPKLDVLDFWKIISPELPTMRGLCTQVTAFMDAP
MAB_4622c|M.abscessus_ATCC_199 MIPSMIVEDGDRPLEMKPKLDVVDYWSIIRAELPAVRGLCTQVTSFSDET
*:*.:* *:.***: *: *:**:::*.:: .*:. :.*******:* .
Mflv_1061|M.gilvum_PYR-GCK TLRGRLALLGDSGFEGIAFVGVPRTLNDGEGTGVAPTDALSIFDGVVENR
Mvan_5770|M.vanbaalenii_PYR-1 TLRGRLTELRDSGFEGIAFVGVPRTLSDGEGSGVAPTDALSIFDQVVENR
MSMEG_6596|M.smegmatis_MC2_155 ALRERLTALSGAGFDGISFVGVPRTMKDGEGEGVAPTDALSIYADLVPNR
MMAR_3207|M.marinum_M ALRRRLVDLSDAGMEGVVFVGVPRTMNDGEGSGVAPTDALSIYRDLLPNR
MUL_3515|M.ulcerans_Agy99 ALRRRLVDLSDAGMEGVVFVGVPRTMNDGEGSGVAPTDALSIYRDLLPNR
MLBr_00901|M.leprae_Br4923 SLGRRLADLSAAGMEGVVFVGVPRTMNDGEGSGVAPTDALSMYRELVANR
Mb2194c|M.bovis_AF2122/97 SLHRRLVDLSDAGMEGIVFVGVPRTMQDGEGSGVAPTDALSLYRQLVANR
Rv2172c|M.tuberculosis_H37Rv SLHRRLVDLSDAGMEGIVFVGVPRTMQDGEGSGVAPTDALSLYRQLVANR
MAV_2322|M.avium_104 SLHGRLTELCSAGMEGVVFVGVPRTMNDGEGSGVAPTDALSIYRDLVTNR
TH_1200|M.thermoresistible__bu ALRRRLTELDAAGFDGVVFVGVPRTMKDGEGSGVPPTDALSMFSDVVANR
MAB_4622c|M.abscessus_ATCC_199 SLRGRLTDLTDVGMDGVIFVGVPRTMNDGDGAGIPPTDALSKFDDIVQNR
:* **. * *::*: *******:.**:* *:.****** : :: **
Mflv_1061|M.gilvum_PYR-GCK GVILIPTREGEHGRFNFKCDRGATYGMTQLLYSDAIVGMLTEFADKTDHR
Mvan_5770|M.vanbaalenii_PYR-1 GVILIPTREGEHGRFNFKCDRGATYGMTQLLYSDAIVGFLKEFAASTDHR
MSMEG_6596|M.smegmatis_MC2_155 GAILIPTREGEQGRFNFKCDQGATYGMTQLLYSDAIVGFLTEFAKTTEHR
MMAR_3207|M.marinum_M GVIVIPTRDGEQGRLNFKCNQGATYGMTQLLYSDAIVKFLTEFADSTDHR
MUL_3515|M.ulcerans_Agy99 GVIVIPTRDGEQGRLNFKCNQGATYGMTQLLYSDATVKFLTEFAYSTDHR
MLBr_00901|M.leprae_Br4923 GVIVIPTRDGEQDRLNFKCNQGATYGMTQLLYSDAIVRFLTEFAKHTDHR
Mb2194c|M.bovis_AF2122/97 GVIVIPTRDGEQGRLNFKCSRGATYGMTQLLYSDAIVGFLREFARTTEHR
Rv2172c|M.tuberculosis_H37Rv GVIVIPTRDGEQGRLNFKCSRGATYGMTQLLYSDAIVGFLREFARTTEHR
MAV_2322|M.avium_104 GVILIPTRDGEQGRFSFKCDRGATYGMTQLLYSDAIVGFLREFARQTDHR
TH_1200|M.thermoresistible__bu GAILIPTRDGEAGRFRFKCERGAMFAMTQLLYSDAIVGFLREFARTTDHR
MAB_4622c|M.abscessus_ATCC_199 GAILIPTRAGEQGRFSFKCDKGANFAMTQLLYSDAVVGFLHEFAHTSDHR
*.*:**** ** .*: ***.:** :.********* * :* *** ::**
Mflv_1061|M.gilvum_PYR-GCK PEILLSFGFVPKVESRVGLINWLIQDPGNEAVAREQEFVRRLADTEPAPR
Mvan_5770|M.vanbaalenii_PYR-1 PEILLSFGFVPKVENRVGLINWLIQDPGNEAVAREQEFVRTLSDTDPAGR
MSMEG_6596|M.smegmatis_MC2_155 PEILLSFGFVPKMEAKVGLINWLIQDPGNPAVAAEQEFVSHLAEQDPAEK
MMAR_3207|M.marinum_M PELLLSFGFIPKVESRVGLINWLIQDPGNPAVAAEQEFVKRLAASEPAQK
MUL_3515|M.ulcerans_Agy99 PELLLSFGFIPKVESRVGLINWLIQDPGNPAVAAEQEFVKRLAASEPAQK
MLBr_00901|M.leprae_Br4923 PELLLSFGFVPKAERRVGLINWLIQDSGNAAVADEQEFVKRLAGSEPAQK
Mb2194c|M.bovis_AF2122/97 PEILLSFGFVPKVETRIGLINWLIQDPGNAAVADEQAFVQKLAGSEPARR
Rv2172c|M.tuberculosis_H37Rv PEILLSFGFVPKVETRIGLINWLIQDPGNAAVADEQAFVQKLAGSEPARR
MAV_2322|M.avium_104 PEILLSFGFVPKMESRVGLINWLIQDPGNAAVAREQEFVKTLAASEPAQK
TH_1200|M.thermoresistible__bu PEILLSFGFVPAVESRMGLINWLIQDPGNPVVAEEQAFVKKLAAAEPDRR
MAB_4622c|M.abscessus_ATCC_199 PEVLLSFGFVPKLESKIGLIDWLIQDPGNEAVAREQDFVKALAAREPGER
**:******:* * ::***:*****.** .** ** ** *: :* :
Mflv_1061|M.gilvum_PYR-GCK RQMMVDLYKRVIDGVADLGFPLSVHFEATYGVNTAAFETLAEMLAYWAPD
Mvan_5770|M.vanbaalenii_PYR-1 RRLMTDLYKRVIDGVADLGFPLSVHFESTYGVNTAAFETLAEMLAYWSPD
MSMEG_6596|M.smegmatis_MC2_155 RRLMVDLYKRVIDGVGQLGFPLSVHYEVAYGVSRPAFETLAEMLEYWSPD
MMAR_3207|M.marinum_M RQLMVDLYKRIMDGVVGLGFPLSIHFEATYGVSPAAFQTFAEMLAYWAPD
MUL_3515|M.ulcerans_Agy99 RQLMVDLYKRIMDGVVGLGFPLSIHFEATYGVSPAAFQTFAEMLAYWAPD
MLBr_00901|M.leprae_Br4923 RQLMLDLYQRVIDGVAELGFPLSIHFEATYGMSPAAFDTFAEMVAYWPPV
Mb2194c|M.bovis_AF2122/97 RRLMVDLYKRVLDGVADLGFPLSIHLEATYGVSAAAFETFAEMLAYWSPA
Rv2172c|M.tuberculosis_H37Rv RRLMVDLYKRVLDGVADLGFPLSIHLEATYGVSAAAFETFAEMLAYWSPA
MAV_2322|M.avium_104 RRQMVDLYKRVIDGVGDLGFPLSIHLEATYGISGPAFQTFAEMLAYWSPR
TH_1200|M.thermoresistible__bu RQLMMDLYKRVIDGVVDLGFPLSIHLEATYGMSAGAFETFAEMLAYWSPE
MAB_4622c|M.abscessus_ATCC_199 RPQLVDLYKRVVDGVGGLGFPLSIHFEAPYGLARPAFETFAEMLEYWSPA
* : ***:*::*** ******:* * .**: **:*:***: **.*
Mflv_1061|M.gilvum_PYR-GCK KP-------
Mvan_5770|M.vanbaalenii_PYR-1 KP-------
MSMEG_6596|M.smegmatis_MC2_155 QG-------
MMAR_3207|M.marinum_M THG------
MUL_3515|M.ulcerans_Agy99 THG------
MLBr_00901|M.leprae_Br4923 CVGQSGEST
Mb2194c|M.bovis_AF2122/97 EPGKPD---
Rv2172c|M.tuberculosis_H37Rv EPGKPD---
MAV_2322|M.avium_104 PS-------
TH_1200|M.thermoresistible__bu TAVSENKKG
MAB_4622c|M.abscessus_ATCC_199 VS-------