For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSFFLRVEVGGLMMAAGRLERITSESMACNAKLTPVTTKVVPPAADQVSKLASQVFSSYGKQYEGYAAQG VDQSRLFVQSLKDAAGDYMDSDHMYLNTED
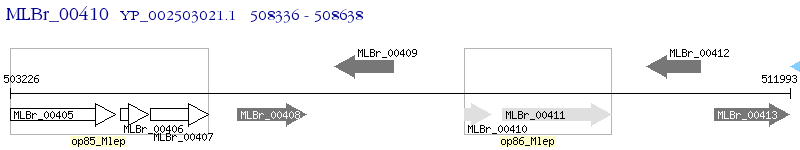
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00410 | - | - | 100% (100) | putative PE-family protein |
| M. leprae Br4923 | MLBr_01183 | - | 5e-14 | 36.96% (92) | hypothetical protein MLBr_01183 |
| M. leprae Br4923 | MLBr_01053 | - | 5e-14 | 36.96% (92) | hypothetical protein MLBr_01053 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2131 | PE22 | 3e-13 | 44.71% (85) | hypothetical protein Mb2131 |
| M. gilvum PYR-GCK | Mflv_0326 | - | 1e-09 | 34.44% (90) | PE domain-containing protein |
| M. tuberculosis H37Rv | Rv2107 | PE22 | 3e-13 | 44.71% (85) | PE family protein |
| M. abscessus ATCC 19977 | MAB_2231c | - | 4e-08 | 34.09% (88) | PE family protein |
| M. marinum M | MMAR_1949 | - | 3e-10 | 39.53% (86) | PE family protein |
| M. avium 104 | MAV_4868 | - | 2e-09 | 35.23% (88) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0618 | - | 3e-08 | 40.58% (69) | PE family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2246 | - | 4e-09 | 39.13% (69) | PE family protein |
| M. vanbaalenii PYR-1 | Mvan_0414 | - | 6e-10 | 36.67% (90) | PE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0326|M.gilvum_PYR-GCK MT--LRVVPEGLTAASAAVEALTARMAAAH-AAAAPLVTAVLPPAADAVS
Mvan_0414|M.vanbaalenii_PYR-1 MT--LRVVPEGLTAAGAAVEALTARMAAAH-AAAAPVVTAVLPPAADVVS
MSMEG_0618|M.smegmatis_MC2_155 MT--LRVVPEGLTAASSAVEALTARLAAAH-AAAAPMISTVLPPAADAVS
MAB_2231c|M.abscessus_ATCC_199 MN--LNVVPEGLTATSAAVEALTARLAAAH-AAAAPVIGAVVPPAADPVS
MAV_4868|M.avium_104 MT--LRVVPEGLAATSAAVEALTARLAAAH-AAAAPAITAVVPPAADPVS
MUL_2246|M.ulcerans_Agy99 MV--LRVVPEGLVATSAAVEALTARLAAAN-AALAPLITAVVPPAADPVS
Mb2131|M.bovis_AF2122/97 MS-FVNVDPFGMLAAAATLESLGSHMAVSN-AAVASVTTKVPPPAADYVS
Rv2107|M.tuberculosis_H37Rv MS-FVNVDPFGMLAAAATLESLGSHMAVSN-AAVASVTTKVPPPAADYVS
MLBr_00410|M.leprae_Br4923 MSFFLRVEVGGLMMAAGRLERITSESMACN-AKLTPVTTKVVPPAADQVS
MMAR_1949|M.marinum_M MS-FLTTQPEELAAAAGKLETIGATMDAENVAAVAPTTTGVIPAAADEVS
* : . : :.. :* : : . : * :. * *.*** **
Mflv_0326|M.gilvum_PYR-GCK LQTATGLSANGVEHQSIAARGVEELGRSGVGVGDSAASYVSGDAMAATSY
Mvan_0414|M.vanbaalenii_PYR-1 LQAATGFSARAAEHQSVAAQGVEELGRSGIAVGESAASYAGGDAMAASSY
MSMEG_0618|M.smegmatis_MC2_155 LQTAAGFSANGAQQSAVAAQGVEELGRSGVGVGESGVSYATGDAQAAASY
MAB_2231c|M.abscessus_ATCC_199 LETAAGFSARGIEHSGVAAQAVEELGRAGLGVSESAASYTTGDMQAAAAY
MAV_4868|M.avium_104 LQTALGFSAQGQEHSAVAAQGVEELGRAGVGVGEAGASYLAGDTAAAATF
MUL_2246|M.ulcerans_Agy99 LEAAIGFSAHGVEHVAVTTEGIEELGRSGVGVGESGLSYASGDAAAALTY
Mb2131|M.bovis_AF2122/97 KKLSLFFSSHGQQYQVQAARGTAFHRKLVRTLANGALAYEEVEIA--NNE
Rv2107|M.tuberculosis_H37Rv KKLSLFFSSHGQQYQVQAARGTAFHRKLVRTLANGALAYEEVEIA--NNE
MLBr_00410|M.leprae_Br4923 KLASQVFSSYGKQYEGYAAQGVDQSRLFVQSLKDAAGDYMDSDHMYLNTE
MMAR_1949|M.marinum_M ALQASLFTAYGTLYQQVSAEAATLYDMFVRTLGVSAGTYAAAETANSSAA
: ::: . ::.. : .. * :
Mflv_0326|M.gilvum_PYR-GCK LIARG---------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 LIARG---------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 LTARGL--------------------------------------------
MAB_2231c|M.abscessus_ATCC_199 MIARG---------------------------------------------
MAV_4868|M.avium_104 GIAGA---------------------------------------------
MUL_2246|M.ulcerans_Agy99 GLFM----------------------------------------------
Mb2131|M.bovis_AF2122/97 GF------------------------------------------------
Rv2107|M.tuberculosis_H37Rv GF------------------------------------------------
MLBr_00410|M.leprae_Br4923 D-------------------------------------------------
MMAR_1949|M.marinum_M ASPLSGIIDALGASTPASDIANIVNIGAGNWASASSDLLALAGGGLLPAA
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2231c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4868|M.avium_104 --------------------------------------------------
MUL_2246|M.ulcerans_Agy99 --------------------------------------------------
Mb2131|M.bovis_AF2122/97 --------------------------------------------------
Rv2107|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00410|M.leprae_Br4923 --------------------------------------------------
MMAR_1949|M.marinum_M VQAAEGDVAQAAGSAESAAAAAGAASLGEAPIVAGVGGVSAMSAASASPS
Mflv_0326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0414|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2231c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_4868|M.avium_104 --------------------------------------------------
MUL_2246|M.ulcerans_Agy99 --------------------------------------------------
Mb2131|M.bovis_AF2122/97 --------------------------------------------------
Rv2107|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00410|M.leprae_Br4923 --------------------------------------------------
MMAR_1949|M.marinum_M WAGQATLVSSTSAGALRGAGWTVPAPQAAPGTVIPGMPGLAAATRNSAGF
Mflv_0326|M.gilvum_PYR-GCK ------------------
Mvan_0414|M.vanbaalenii_PYR-1 ------------------
MSMEG_0618|M.smegmatis_MC2_155 ------------------
MAB_2231c|M.abscessus_ATCC_199 ------------------
MAV_4868|M.avium_104 ------------------
MUL_2246|M.ulcerans_Agy99 ------------------
Mb2131|M.bovis_AF2122/97 ------------------
Rv2107|M.tuberculosis_H37Rv ------------------
MLBr_00410|M.leprae_Br4923 ------------------
MMAR_1949|M.marinum_M GAPRYGVKPIVMPKPAAV