For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LIAAIPPEPTSTYLVLLQIGMYGMMLLTGVPVLIQAVLPELLRRDFDHDLVHQPETMNLFVELFVPYLLA TGYTGSLSPTTR
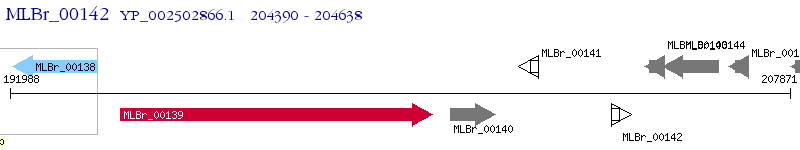
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00142 | - | - | 100% (82) | hypothetical protein MLBr_00142 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1403 | - | 2e-16 | 59.70% (67) | peptidase M1, membrane alanine aminopeptidase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0509c | - | 3e-13 | 47.06% (68) | peptidase M1, membrane alanine aminopeptidase |
| M. marinum M | MMAR_5284 | pepN_1 | 1e-18 | 58.82% (68) | aminopeptidase N PepN_1 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6138 | - | 3e-17 | 61.19% (67) | metallopeptidase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4357 | pepN_1 | 8e-19 | 58.82% (68) | aminopeptidase N PepN_1 |
| M. vanbaalenii PYR-1 | Mvan_5387 | - | 6e-16 | 59.70% (67) | peptidase M1, membrane alanine aminopeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0509c|M.abscessus_ATCC_199 MTKPGKKAAKRLPPRPPVIDPYLPQNGNYGYRVSRYELDLEYKVAINRLT
MSMEG_6138|M.smegmatis_MC2_155 MTRS-KKVAKKAG-HPPVIDPYLPGNGNFGYRVSRYELDLEYKVASNRLT
Mflv_1403|M.gilvum_PYR-GCK -------MSKKNS--SPVIDPYLPEAGNFGYRVSRYELDLEYKTTINRLS
Mvan_5387|M.vanbaalenii_PYR-1 MTAR-KRAAKKSS--PPVIDPYLPGAGNFGYRVSRYELELEYKVTINRLS
MMAR_5284|M.marinum_M MKASAKMPKKAIK----IIDPYLPQNGNFGYRVSRYELDLEYKVAINRLS
MUL_4357|M.ulcerans_Agy99 MKASAKMPKKAIK----IIDPYLPQNGNFGYRVSRYELDLEYKVAINRLS
MLBr_00142|M.leprae_Br4923 --------------------------------------------------
MAB_0509c|M.abscessus_ATCC_199 GTANITAVTLATIKEFTLDLADTLGVAKVTVNGRRPTHFGHRAGKITIKL
MSMEG_6138|M.smegmatis_MC2_155 GTATITAVTLAALRTFTLDLSDNLSVGKVSVNGRRPAQFRCSGAKLHITL
Mflv_1403|M.gilvum_PYR-GCK GSAAITAVTLAEIKTFTLDLADALSVSKVSVNGKRPAHYRTAAGKLHITL
Mvan_5387|M.vanbaalenii_PYR-1 GAATITAVTLAELKTFTLDLSGALSVSKVTVNGRRPAHYRTSADKLHITL
MMAR_5284|M.marinum_M GTATITAVTLTELQEFTLDLSDALSVSKVLVNGKRVARFASRNGKLRIGL
MUL_4357|M.ulcerans_Agy99 GTATITAVTLTELQEFTLDLSDALSVLKVLVNGKRVAHFASRNGKLRIGL
MLBr_00142|M.leprae_Br4923 --------------------------------------------------
MAB_0509c|M.abscessus_ATCC_199 ASAIPAGAAMTIVIRYGGSPRPINSMWGDVGFEELSNGVLVAGQPNGAAS
MSMEG_6138|M.smegmatis_MC2_155 PAALPAGSAMTVVIRYSGNPRPIDTLWGDVGFEELTNGALVAGQPNGAAS
Mflv_1403|M.gilvum_PYR-GCK AERLPAGAAMTVGIRYSGNPRPNRTPWGDVGFEELTEGVLVAGQPNGASS
Mvan_5387|M.vanbaalenii_PYR-1 SERLPAGAAMTIAVRYGGTPRPIRSLWGEVGFEELTDGVLVAGQPNGASS
MMAR_5284|M.marinum_M TSKLATGVAFSVLVRYSGAPRPLQTLWGEVGFEELTEGALVAGQPNGAAS
MUL_4357|M.ulcerans_Agy99 TSKLATGVAFSVLVRYSGAPRPLQTLWGEVGFEELTEGALVAGQPNGAAS
MLBr_00142|M.leprae_Br4923 ---------------------------------------MIAAIP-----
::*. *
MAB_0509c|M.abscessus_ATCC_199 WFPCDDHPSSKASYRFVITSDSPYCVISNGELISRRVRAAHTTWTYEQPE
MSMEG_6138|M.smegmatis_MC2_155 WFPCDDHPSAKASYRIQISTDSPYHAVANGELLSRKVRAGHTVWTYELAE
Mflv_1403|M.gilvum_PYR-GCK WFPCDDHPAAKASFRVQIATESPYFALANGKLLSRRPRAGMTTWVYEQSE
Mvan_5387|M.vanbaalenii_PYR-1 WFPCDDHPSAKASFRVQIATESAYFALANGKLLTRRARAGMTTWIYEQAE
MMAR_5284|M.marinum_M WFPCNDHPSAKAAFRIQISTESSYRAIANGKLVGQRVRASQTVWTYEQPE
MUL_4357|M.ulcerans_Agy99 WFPCNDHPSAKAAFRIQISTESSYRAIANGKLVGQRVRASPTVWTYEQPE
MLBr_00142|M.leprae_Br4923 ------------------------------------------------PE
.*
MAB_0509c|M.abscessus_ATCC_199 PMAPYLATLQIGDYQLVKLAKNPISMNAALPSRLRDRFEHDFGRQPQMMK
MSMEG_6138|M.smegmatis_MC2_155 PTSPYLVTLQIGMYESHRMTKTPVPMHAVLPPRLRSAFDHDFERQPQMMK
Mflv_1403|M.gilvum_PYR-GCK PTSTYLITLQIGVYNRHRLSKNGVEIFAVLPDRLRREFAHDFGRQPQMMK
Mvan_5387|M.vanbaalenii_PYR-1 PTSTYLITLQIGMYERHRMAKNGVEIFAVLPRRLRDDFDHDFGRQPQMMK
MMAR_5284|M.marinum_M PTSTYLITLQVGVYVMARMSRSPVPIRAALPERLRSEFDHDFAGQPAMMQ
MUL_4357|M.ulcerans_Agy99 PTSTYLITLQVGVYVMARMSRSPVPIRAALPERLRSEFDHDFAGQPAMMQ
MLBr_00142|M.leprae_Br4923 PTSTYLVLLQIGMYGMMLLTGVPVLIQAVLPELLRRDFDHDLVHQPETMN
* :.** **:* * :: : : *.** ** * **: ** *:
MAB_0509c|M.abscessus_ATCC_199 LFIKQFGPYPFAAGYTVVVTDDDLDIPLEAQGISVFGANHCDGSRHSERL
MSMEG_6138|M.smegmatis_MC2_155 LFVKLFGPYPLSTGYTVVVTDDDLEIPLEAQGISIFGANHCDGSRGSERL
Mflv_1403|M.gilvum_PYR-GCK LFTKLFGPYPLATGYTVVVTDDDLEIPLEAQGISIFGANHCDGSRKAERL
Mvan_5387|M.vanbaalenii_PYR-1 LFTKLFGPYPLANGYTVVVTDDDLEIPLEAQGISIFGANHCDGRRGEERL
MMAR_5284|M.marinum_M LFIDLFGPYPLANGYTVVVTDDALEIPLEAQGISIFGANHCDGTRSSERL
MUL_4357|M.ulcerans_Agy99 LFIDLFGPYPLANGYTVVVTDDALEIPLEAQGISIFGANHCDGTRSSERL
MLBr_00142|M.leprae_Br4923 LFVELFVPYLLATGYT--------------------------GS------
** . * ** :: *** *
MAB_0509c|M.abscessus_ATCC_199 VAHELAHQWFGNSVTARRWRDIWLHEGFACYAEWLWSEESGGPTADARAR
MSMEG_6138|M.smegmatis_MC2_155 IAHELAHQWFGNSVTARRWRDIWLHEGFACYAEWLWSENSGGRGAAEWAQ
Mflv_1403|M.gilvum_PYR-GCK IAHELAHQWFGNSVTVQRWRHIWLHEGFACYAEWLWSEHCGERTAQQWAQ
Mvan_5387|M.vanbaalenii_PYR-1 IAHELAHQWFGNSVTVQRWRHIWLHEGFACYAEWLWSEHCGERTVDQWAR
MMAR_5284|M.marinum_M IAHELAHQWFGNSVTARRWCDIWLHEGFACYAEWLWSEHSGGPSADELAR
MUL_4357|M.ulcerans_Agy99 IAHELAHQWFGNSVTARRWCDIWLHEGFACYAEWLWSEHSGGPSADELAR
MLBr_00142|M.leprae_Br4923 -----------LSPTTR---------------------------------
* *.:
MAB_0509c|M.abscessus_ATCC_199 HYHQKLKDLPQDLILADPGPQLMFDDRVYKRGALTLHALRLRLGDTKFFA
MSMEG_6138|M.smegmatis_MC2_155 HYYDKLRHSPQDLLLADPGPRDMFDDRVYKRGALTLHVLRERIGDKNFFA
Mflv_1403|M.gilvum_PYR-GCK HYHHRLKHSPHDLVLADPGPRDMFDDRVYKRGALTLHVLRQRIGDANFFA
Mvan_5387|M.vanbaalenii_PYR-1 HYHERLKNSPHNLVLADPGPRDMFDDRVYKRGALTLHALRNHIGDTEFFA
MMAR_5284|M.marinum_M YHHQELRGKPQDLLLTDPGPQLMFDDRVYKRGALALHSLRGRLGDAKFFA
MUL_4357|M.ulcerans_Agy99 YHHQELRGKPQDLLLTDPGPQLMFDDRVYKRGALALHSLRGRLGDAKFFA
MLBr_00142|M.leprae_Br4923 --------------------------------------------------
MAB_0509c|M.abscessus_ATCC_199 LLQDWTTRYRHSTVITDDFTGLAAGYSDESLRPLWTAWLYEKQLPPLDRK
MSMEG_6138|M.smegmatis_MC2_155 LLQDWTTRHRHGTAFTDDFTGLAAGYTSESLQPLWQAWLYSEALPPL---
Mflv_1403|M.gilvum_PYR-GCK LLKDWTTRYRHSSVVTDDFLGLASQYSDESLRSLWAAWLYSTEVPALDAG
Mvan_5387|M.vanbaalenii_PYR-1 LLKDWTNRYRHSSVVTDDFVGLAARYAHDSLRPLWAAWLYSPEVPELDAG
MMAR_5284|M.marinum_M LLRDWTTRYRHGTVITDDFTGLAANYATESLRDLWDTWLYSTAVP-----
MUL_4357|M.ulcerans_Agy99 LLRDWTTRYRHGTVITDDFTGLAANYATESLRDLWDTWLYSTAVP-----
MLBr_00142|M.leprae_Br4923 --------------------------------------------------
MAB_0509c|M.abscessus_ATCC_199 -
MSMEG_6138|M.smegmatis_MC2_155 -
Mflv_1403|M.gilvum_PYR-GCK S
Mvan_5387|M.vanbaalenii_PYR-1 S
MMAR_5284|M.marinum_M -
MUL_4357|M.ulcerans_Agy99 -
MLBr_00142|M.leprae_Br4923 -