For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAKPPTIIYTLTDEAPLLATYALLPVVQTFAAAAGINVKTSDISLAARILAEFPDHLTEDQRVPNNLAEL AELTQLPDTNIIKLPNVSASVPQLLAAIKELQAKGFDLPNYPGDPKTDEEKEITQRYARCLGSAVNPVLR EGNSDRRAPKPVKQYARKHPHSMGEWSMASRTHVATMKHGDFYHGEKSMTLDRARGVKMELKTKGGKTIV LKPKVSLRDGDVIDSMFMSKKALCEFYEAQMQDAYETGVMFSLHVKATMMKVSHPIVFGHAVKIFYKDAF AKHQELFDELGVNVNNGLVDLYSKIETLPASLHDEIVRDLHACHEHRPELAMVDSAKGITNFHSPSDVIV DASMPAMIRLGGKMYGADGRTKDTKAVSPESTFSRIYQEIINFCKTHGQFDPTTMGTVPNVGLMAQQAEE YGSHDKTFEIPEDGVANIVDVDTGEVLLTQDVETGDIWRMPIVRDDPIRDWVKLAVDRARNSGMPVVFWL DQERPHENELRKKVNTYLADQDTEGLDIQTMSQERAMRHTIERAMRGQDTIAATGNILRDYLTDLFPILE LGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVHQLVEENHLRWDSLGEYLALGACFEDTGIKTGNKRATI LAKSLDAATGKLLDNNKSPSRKTGELDNRGSQFYLALYWAQELAAQHDDEELRKHFAALADELRNHEDTI LAELAKAQGESVDIGGYYRPDDEKLTAVMRPSKTLNDALVTHRADAGRAKLVQ*
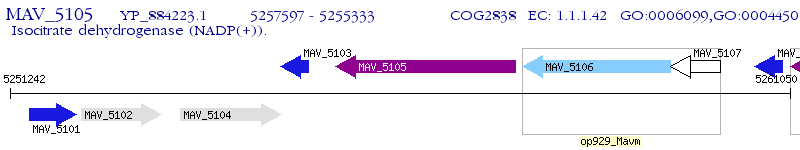
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5105 | - | - | 100% (754) | isocitrate dehydrogenase, NADP-dependent |
| M. avium 104 | MAV_4313 | - | 0.0 | 84.73% (740) | isocitrate dehydrogenase, NADP-dependent |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0067c | icd2 | 0.0 | 83.11% (740) | isocitrate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3455 | - | 0.0 | 79.17% (749) | isocitrate dehydrogenase, NADP-dependent |
| M. tuberculosis H37Rv | Rv0066c | icd2 | 0.0 | 83.24% (740) | isocitrate dehydrogenase |
| M. leprae Br4923 | MLBr_02672 | icd2 | 0.0 | 82.03% (740) | isocitrate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3686c | - | 0.0 | 79.65% (742) | isocitrate dehydrogenase |
| M. marinum M | MMAR_0158 | icd2 | 0.0 | 83.78% (740) | isocitrate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1654 | - | 0.0 | 80.68% (740) | isocitrate dehydrogenase, NADP-dependent |
| M. thermoresistible (build 8) | TH_1971 | icd2 | 0.0 | 81.08% (740) | PROBABLE ISOCITRATE DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_4939 | icd2 | 0.0 | 83.51% (740) | isocitrate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3212 | - | 0.0 | 80.84% (741) | isocitrate dehydrogenase, NADP-dependent |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1654|M.smegmatis_MC2_155 MSAQQPTIIYTLTDEAPLLATYAFLPVVRKFAEAAGIDVKTSDISVAARI
TH_1971|M.thermoresistible__bu MSAEKPTIIYTLTDEAPLLATYSFLPIIESFAKPAGIDIKTSDISVAARI
Mflv_3455|M.gilvum_PYR-GCK MSAEQPSIIYTLTDEAPLLATYAFLPVVRTFASAAGIDVKSTDISVAARI
Mvan_3212|M.vanbaalenii_PYR-1 MSAEQPSIIYTLTDEAPLLATYAFLPVVRTFASAAGIDVKSSDISVAARI
MAB_3686c|M.abscessus_ATCC_199 MSAQQPTIIYTLTDEAPLLATYSFLPIIRAFAGPAGIDVQTSDISVAARI
Mb0067c|M.bovis_AF2122/97 MSAEQPTIIYTLTDEAPLLATYAFLPIVRAFAEPAGIKIEASDISVAARI
Rv0066c|M.tuberculosis_H37Rv MSAEQPTIIYTLTDEAPLLATYAFLPIVRAFAEPAGIKIEASDISVAARI
MMAR_0158|M.marinum_M MSAEQPTIIYTLTDEAPLLATYAFLPIVRAFAEPAGIQIKTSDISVAARI
MUL_4939|M.ulcerans_Agy99 MSAEQPTIIYTLTDEAPLLATYAFLPIVRAFAEPAGIQIKTSDVSVAARI
MLBr_02672|M.leprae_Br4923 MSAEQWTIIYTLTDEAPLLATYAFLPIVRAFTKPAGIKVQTSDISVAARI
MAV_5105|M.avium_104 MSAKPPTIIYTLTDEAPLLATYALLPVVQTFAAAAGINVKTSDISLAARI
***: :***************::**::. *: .***.::::*:*:****
MSMEG_1654|M.smegmatis_MC2_155 LAEFGDHLTEEQRVPDNLGELGALTQDPSANIIKLPNISASVPQLLAAIK
TH_1971|M.thermoresistible__bu LAEFPDYLTEEQRVPDNLGELGRLTQDPDTNIIKLPNISASVPQLLAAIK
Mflv_3455|M.gilvum_PYR-GCK LAEFSDRLTDEQKVTDNLAELGELTQLPETNIIKLPNISASVPQLLAAIK
Mvan_3212|M.vanbaalenii_PYR-1 LAEFSDRLTDEQKVPDNLAELGALTQEPDTNIIKLPNISASVPQLLAAIK
MAB_3686c|M.abscessus_ATCC_199 LSEFSEYLPEDQRVPDNLAELGRLTQLPETNIIKLPNISASVPQLLAAIK
Mb0067c|M.bovis_AF2122/97 LAEFPDYLTEEQRVPDNLAELGRLTQLPDTNIIKLPNISASVPQLVAAIK
Rv0066c|M.tuberculosis_H37Rv LAEFPDYLTEEQRVPDNLAELGRLTQLPDTNIIKLPNISASVPQLVAAIK
MMAR_0158|M.marinum_M LAEFPDYLTDEQRVPDNLAELGELTQDPDTNIIKLPNISASVPQLKAAIK
MUL_4939|M.ulcerans_Agy99 LAEFPDYLTDEQRVPDNLAELGELTQDPDTNIIKLPNISASVPQLKAAIK
MLBr_02672|M.leprae_Br4923 LAEFPDYLTEEQRVPNNLAELGRLTQLPDTNIIKLPNISASVPQLMAAIK
MAV_5105|M.avium_104 LAEFPDHLTEDQRVPNNLAELAELTQLPDTNIIKLPNVSASVPQLLAAIK
*:** : *.::*:*.:**.**. *** *.:*******:******* ****
MSMEG_1654|M.smegmatis_MC2_155 ELQGKGYNVPDYPANPKTDDEKKIKDRYAKILGSAVNPVLREGNSDRRAP
TH_1971|M.thermoresistible__bu ELQGKGFNVPDYPGDPKTEEEQKIKDRYAKVLGSAVNPVLREGNSDRRAP
Mflv_3455|M.gilvum_PYR-GCK ELKAKGYDLPDWVGEPKNDEEREIKERYSKVLGSAVNPVLRQGNSDRRAP
Mvan_3212|M.vanbaalenii_PYR-1 ELKAKGYDLPDYPGDPKTDEEKAVKERYAKVLGSAVNPVLRQGNSDRRAP
MAB_3686c|M.abscessus_ATCC_199 ELQGKGYKLPEYPGDPKNEEETEIKARYTKCLGSAVNPILREGNSDRRAP
Mb0067c|M.bovis_AF2122/97 ELQDKGYAVPDYPADPNTDQEKAIKERYARCLGSAVNPVLRQGNSDRRAP
Rv0066c|M.tuberculosis_H37Rv ELQDKGYAVPDYPADPKTDQEKAIKERYARCLGSAVNPVLRQGNSDRRAP
MMAR_0158|M.marinum_M ELQDKGYAIPDYPADPKTDEEKALKERYARCLGSAVNPVLRQGNSDRRAP
MUL_4939|M.ulcerans_Agy99 ELQDKGYAIPDYPADPKTDEEKALKERYARCLGSAVNPVLRQGNSDRRAP
MLBr_02672|M.leprae_Br4923 ELQAKGFKLPDYPQDPKSYQEIDIRNRYGKCLGSAVNPVLREGNSDRRAP
MAV_5105|M.avium_104 ELQAKGFDLPNYPGDPKTDEEKEITQRYARCLGSAVNPVLREGNSDRRAP
**: **: :*:: :*:. :* : ** : *******:**:********
MSMEG_1654|M.smegmatis_MC2_155 KAVKEYARKHPHSMGEWSQASRTHVATMKTGDFYHGEKSMTLDRDRRVKM
TH_1971|M.thermoresistible__bu KAVKEYARKNPHSMGEWSQASRTHVATMKKGDFYAGEKSMTLDRARRVKM
Mflv_3455|M.gilvum_PYR-GCK KAVKEYARKHPHSMGEWSQASRTHVATMKTGDFYHGEKSMTVDKDRKVKM
Mvan_3212|M.vanbaalenii_PYR-1 KAVKEYARKHPHSMGEWSQASRTHVATMKHGDFYHGEKSMTLDKDRKVKM
MAB_3686c|M.abscessus_ATCC_199 KAVKEYARKHPHSMGEWSQASRTHVATMKEGDFYHGEKSLTLDRDRTVKM
Mb0067c|M.bovis_AF2122/97 KAVKEYARKHPHSMGEWSMASRTHVAHMRHGDFYAGEKSMTLDRARNVRM
Rv0066c|M.tuberculosis_H37Rv KAVKEYARKHPHSMGEWSMASRTHVAHMRHGDFYAGEKSMTLDRARNVRM
MMAR_0158|M.marinum_M NAVKEYARKHPHSMGEWSMASRTHVAHMRHGDFYAGEKSITLDRAHKVKM
MUL_4939|M.ulcerans_Agy99 NAVKEYARKHPHSMGEWSMASRTHVAHMRHGDFYAGEKSITLDRAHKVKM
MLBr_02672|M.leprae_Br4923 KAVKEYARNNPHSMGEWSMASRTHVATMRHGDFYHGEKSMTLDRARDVKM
MAV_5105|M.avium_104 KPVKQYARKHPHSMGEWSMASRTHVATMKHGDFYHGEKSMTLDRARGVKM
:.**:***::******** ******* *: **** ****:*:*: : *:*
MSMEG_1654|M.smegmatis_MC2_155 VLK----TKSGEEIVLKPEVKLDAGDIIDSMYMSKKALIAFYEEQIEDAY
TH_1971|M.thermoresistible__bu VLN----TKRGETLVLKPEVKLDEGDIIDSMFMSKKALCEFYEEQMQDAY
Mflv_3455|M.gilvum_PYR-GCK VLTTERPDGSSETQVLKPEVSLDEGDVIDSMFMSKKALVEFYEEQIEDAY
Mvan_3212|M.vanbaalenii_PYR-1 VLE---PSGGGPALVLKPEVALEKGDVIDSMYMSKKALIDFYEEQIEDAY
MAB_3686c|M.abscessus_ATCC_199 VLT----TKSGDTVVLKPEVKLGKGDIIDSMFMSKKALCDFYEEQIEDAY
Mb0067c|M.bovis_AF2122/97 ELL----AKSGKTIVLKPEVPLDDGDVIDSMFMSKKALCDFYEEQMQDAF
Rv0066c|M.tuberculosis_H37Rv ELL----AKSGKTIVLKPEVPLDDGDVIDSMFMSKKALCDFYEEQMQDAF
MMAR_0158|M.marinum_M ELV----TKGGQTLVLKPEVSLDDGDIIDSMFMSKKALCEFYEEQIEDAY
MUL_4939|M.ulcerans_Agy99 ELV----TKGGQTLVLKPEVSLDDGDIIDSMFMSKKALCEFYEEQIEDAY
MLBr_02672|M.leprae_Br4923 ELA----TKKGKIVVLKPKVSLRDGDIIDSMFMSKKALCEFYEEQMEDAR
MAV_5105|M.avium_104 ELK----TKGGKTIVLKPKVSLRDGDVIDSMFMSKKALCEFYEAQMQDAY
* . ****:* * **:****:****** *** *::**
MSMEG_1654|M.smegmatis_MC2_155 KTGVMFSLHVKATMMKVSHPIVFGHAVKVFYKDAFAKHEKLFDELGVNVN
TH_1971|M.thermoresistible__bu ETGVMFSLHVKATMMKVSHPIVFGHAVKVFYKDAFAKHQKLFDELGVNVN
Mflv_3455|M.gilvum_PYR-GCK KTGVMFSLHVKATMMKVSHPIVFGHAVKVFYKDAFEKHGKLFKELGVNVN
Mvan_3212|M.vanbaalenii_PYR-1 KTGVMFSLHVKATMMKVSHPIVFGHAVKVFYKDAFAKHQKLFDELGVNVN
MAB_3686c|M.abscessus_ATCC_199 KTGVMLSLHVKATMMRVSHPIVFGHAVKIFYKDAFEKHQKLFDELGVNVN
Mb0067c|M.bovis_AF2122/97 ETGVMFSLHVKATMMKVSHPIVFGHAVRIFYKDAFAKHQELFDDLGVNVN
Rv0066c|M.tuberculosis_H37Rv ETGVMFSLHVKATMMKVSHPIVFGHAVRIFYKDAFAKHQELFDDLGVNVN
MMAR_0158|M.marinum_M KTGVMFSLHVKATMMKVSHPIVFGHAVKTFYRDAFAKHQKLFDELGVNVN
MUL_4939|M.ulcerans_Agy99 KTGVMFSLHVKATMMKVSHPIVFGHAVKTFYRDAFAKHQKLFDELGVNVN
MLBr_02672|M.leprae_Br4923 KTGVMFSLHVKATMMKVSHPIVFGHAVKTFYKEAFAKHQDLFDELGVNVN
MAV_5105|M.avium_104 ETGVMFSLHVKATMMKVSHPIVFGHAVKIFYKDAFAKHQELFDELGVNVN
:****:*********:***********: **::** ** .**.:******
MSMEG_1654|M.smegmatis_MC2_155 NGLSDLYDKIEALPASQREEIIEDLHKCHEHRPELAMVDSAKGISNFHSP
TH_1971|M.thermoresistible__bu NGMSDLYEKISTLPASQREEIIDDIHRCHETRPELAMVDSARGITNFHSP
Mflv_3455|M.gilvum_PYR-GCK NGMSDLYDKIESLPASQREEIIEDIHRCHEHRPELAMVDSARGITNFHSP
Mvan_3212|M.vanbaalenii_PYR-1 NGLSDLYSKIEVLPASQREEIIEDLHRCHEHRPELAMVDSAKGISNFHSP
MAB_3686c|M.abscessus_ATCC_199 NGLSDLYSKIEALPASQHEEIIRDLHACHEHRPELAMVDSAKGISNFHSP
Mb0067c|M.bovis_AF2122/97 NGLSDLYSKIESLPASQRDEIIEDLHRCHEHRPELAMVDSARGISNFHSP
Rv0066c|M.tuberculosis_H37Rv NGLSDLYSKIESLPASQRDEIIEDLHRCHEHRPELAMVDSARGISNFHSP
MMAR_0158|M.marinum_M NGLSDLYSKIETLPASQHDEIIDDLHRCHEHRPELAMVDSARGITNFHSP
MUL_4939|M.ulcerans_Agy99 NGLSDLYSKIETLPASQHDEIIDDLHRCHEHRPELAMVDSARGITNFHSP
MLBr_02672|M.leprae_Br4923 NGLVDLYSKIETLTASQRDEIIQDLHACHEHRPELAMVDSARGISNFYSP
MAV_5105|M.avium_104 NGLVDLYSKIETLPASLHDEIVRDLHACHEHRPELAMVDSAKGITNFHSP
**: ***.**. *.** ::**: *:* *** **********:**:**:**
MSMEG_1654|M.smegmatis_MC2_155 SDVIVDASMPAMIRLGGKMYGADGRTKDTKAVNPESTFSRMYQEMINFCK
TH_1971|M.thermoresistible__bu SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEMINFCK
Mflv_3455|M.gilvum_PYR-GCK SDVIVDASMPAMIRLGGKMYGADGRTKDTKAVNPESTFSRIYQEIINFCK
Mvan_3212|M.vanbaalenii_PYR-1 SDVIVDASMPAMIRLGGKMYGADGRTKDTKAVNPESTFSRMYQEVINFCK
MAB_3686c|M.abscessus_ATCC_199 SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEMINFVK
Mb0067c|M.bovis_AF2122/97 SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEIINFCK
Rv0066c|M.tuberculosis_H37Rv SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEIINFCK
MMAR_0158|M.marinum_M SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEIINFCK
MUL_4939|M.ulcerans_Agy99 SDVIVDASMPAMIRAGGKMYGADGKLKDTKAVNPESTFSRIYQEIINFCK
MLBr_02672|M.leprae_Br4923 SDVIVDASMPAMIRAGGKMYGADGRAKDTKAVNPESTFSRIYQEIINFCK
MAV_5105|M.avium_104 SDVIVDASMPAMIRLGGKMYGADGRTKDTKAVSPESTFSRIYQEIINFCK
************** *********: ******.*******:***:*** *
MSMEG_1654|M.smegmatis_MC2_155 THGQFDPTTMGTVPNVGLMAQKAEEYGSHDKTFEIPEDGVADIVDIDTGE
TH_1971|M.thermoresistible__bu THGQFDPTTMGTVPNVGLMAQKAEEYGSHDKTFEIPEDGVADIVDIDTGE
Mflv_3455|M.gilvum_PYR-GCK THGQFDPTTMGTVPNVGLMAMKAEEYGSHDKTFEITEDGVADIVDIETGE
Mvan_3212|M.vanbaalenii_PYR-1 TNGQFDPRTMGTVPNVGLMAMKAEEYGSHDKTFEIPEDGVANIVDIDTGE
MAB_3686c|M.abscessus_ATCC_199 THGQFDPRTMGTVPNVGLMAQKAEEYGSHDKTFEIAEAGVADIVDIDTGE
Mb0067c|M.bovis_AF2122/97 TNGQFDPTTMGTVPNVGLMAQQAEEYGSHDKTFEIPEDGVANIVDVATGE
Rv0066c|M.tuberculosis_H37Rv TNGQFDPTTMGTVPNVGLMAQQAEEYGSHDKTFEIPEDGVANIVDVATGE
MMAR_0158|M.marinum_M TNGQFDPTTMGTVPNVGLMAQQAEEYGSHDKTFEVPEDGVANIVDLDTGE
MUL_4939|M.ulcerans_Agy99 TNGQFDPTTMGTVPNVGLMAQQAEEYGSHDKTFEVPEDGVANIVDLDTGE
MLBr_02672|M.leprae_Br4923 TNGAFDPATMGTVPNVGLMAQQAEEYGSHDKTFEIPEDGVANIVDLTTGE
MAV_5105|M.avium_104 THGQFDPTTMGTVPNVGLMAQQAEEYGSHDKTFEIPEDGVANIVDVDTGE
*:* *** ************ :************:.* ***:***: ***
MSMEG_1654|M.smegmatis_MC2_155 VLLTQNVEEGDIWRMPIVKDAPIRDWVKLAVTRARLSGMPVVFWLDTERP
TH_1971|M.thermoresistible__bu VLMSQNVEEGDIWRMPIVKDAPIQDWVKLAVTRARASGMPVVFWLDIERP
Mflv_3455|M.gilvum_PYR-GCK VLLSQNVEEGDIWRMPIVKDAAIRDWVKLAVNRARQSGMTTVFWLDDERP
Mvan_3212|M.vanbaalenii_PYR-1 VLLTQNVEEGDIWRMPVVKDAAIRDWVKLAVTRARLSGMPAVFWLDDERP
MAB_3686c|M.abscessus_ATCC_199 VLLTQNVEEGDIWRMPVVTDAAIQDWVKLAVTRGRESGMNVVFWLDTERP
Mb0067c|M.bovis_AF2122/97 VLLTENVEAGDIWRMCIVKDAPIRDWVKLAVTRARISGMPVLFWLDPYRP
Rv0066c|M.tuberculosis_H37Rv VLLTENVEAGDIWRMCIVKDAPIRDWVKLAVTRARISGMPVLFWLDPYRP
MMAR_0158|M.marinum_M VLLTQDVEAGDIWRMCVVKDAPIRDWVKLAVTRARNSGMPVLFWLDPYRP
MUL_4939|M.ulcerans_Agy99 VLLTQDVEAGDIWRMCVVKDAPIRDWVKLAVTRARNSGMPVLFWLDPYRP
MLBr_02672|M.leprae_Br4923 VLLTQNVETGDIWRLCVVTDAAIRDWVKLAVSRMRNSGMPVLFWLDPYRP
MAV_5105|M.avium_104 VLLTQDVETGDIWRMPIVRDDPIRDWVKLAVDRARNSGMPVVFWLDQERP
**::::** *****: :* * .*:******* * * *** .:**** **
MSMEG_1654|M.smegmatis_MC2_155 HEVELRKKVKEYLKDHDTEGLKIQIMPQVWAMRYTLERVVRGKDTIAATG
TH_1971|M.thermoresistible__bu HEVELRKKVKKYLKDHDTEGLKIHILPQVWAMRYTLERVIRGKDTIAATG
Mflv_3455|M.gilvum_PYR-GCK HENELRKKVKTYLKEEDTDGLEITILPQVWAMRYTLERVIRGQDTIAVTG
Mvan_3212|M.vanbaalenii_PYR-1 HENELREKVKAYLKEEDTDGLDITILPQVWAMRYTLERVIRGQDTIAVTG
MAB_3686c|M.abscessus_ATCC_199 HEVELRKKVKEYLQDHDTEGLKIQVVPQVWAMRYTLERLIRGKDTIAATG
Mb0067c|M.bovis_AF2122/97 HENELIKKVKTYLKDHDTEGLDIQIMSQVRSMRYTCERLVRGLDTIAATG
Rv0066c|M.tuberculosis_H37Rv HENELIKKVKTYLKDHDTEGLDIQIMSQVRSMRYTCERLVRGLDTIAATG
MMAR_0158|M.marinum_M HENELIKKVKTYLKDHHTEGLDIQIMSQVRSMRYTLERLIRGLDTIAATG
MUL_4939|M.ulcerans_Agy99 HENELIKKVKTYLKDHHTEGLDIQIMSQVRSMRYTLERLIRGLDTIAATG
MLBr_02672|M.leprae_Br4923 HENELIKKVRAYLKDHDTSGLDIQIMSQVRSMRYTCERLIRGLDTIAATG
MAV_5105|M.avium_104 HENELRKKVNTYLADQDTEGLDIQTMSQERAMRHTIERAMRGQDTIAATG
** ** :**. ** :..*.**.* :.* :**:* ** :** ****.**
MSMEG_1654|M.smegmatis_MC2_155 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGLYETGAGGSAPKHVHQL
TH_1971|M.thermoresistible__bu NILRDYLTDLFPILELGTSAKMLSIVPLMKGGGLYETGAGGSAPKHVHQL
Mflv_3455|M.gilvum_PYR-GCK NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVSQL
Mvan_3212|M.vanbaalenii_PYR-1 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVHQL
MAB_3686c|M.abscessus_ATCC_199 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGLYETGAGGSAPKHVSQL
Mb0067c|M.bovis_AF2122/97 NILRDYLTDLFPILELGTSAKMLSVVPLMAGGGMYETGAGGSAPKHVKQL
Rv0066c|M.tuberculosis_H37Rv NILRDYLTDLFPILELGTSAKMLSVVPLMAGGGMYETGAGGSAPKHVKQL
MMAR_0158|M.marinum_M NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVKQL
MUL_4939|M.ulcerans_Agy99 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVKQL
MLBr_02672|M.leprae_Br4923 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVKQL
MAV_5105|M.avium_104 NILRDYLTDLFPILELGTSAKMLSIVPLMAGGGMYETGAGGSAPKHVHQL
************************:**** ***:************* **
MSMEG_1654|M.smegmatis_MC2_155 VEENHLRWDSLGEFLALGASLEDMGNKTGNEKAKVLAKALDTATGKLLEE
TH_1971|M.thermoresistible__bu LEENHLRWDSLGEFLALGACFEDVARKTGNKRADLLGKTLDTAIGKLLDN
Mflv_3455|M.gilvum_PYR-GCK LEENHLRWDSLGEFLALGASFEDMGAKAGNKKAELLGKTLDSAIGKLLDN
Mvan_3212|M.vanbaalenii_PYR-1 VEENHLRWDSLGEFLALGASFEDMGAKFGNKKAELLGKTLDSAIGKLLDN
MAB_3686c|M.abscessus_ATCC_199 VEENHLRWDSLGEFLALAVSLEDLGIKTGNARAKILATTLDAATGKLLEN
Mb0067c|M.bovis_AF2122/97 VEENHLRWDSLGEFLALGAGFEDIGIKTGNERAKLLGKTLDAAIGKLLDN
Rv0066c|M.tuberculosis_H37Rv VEENHLRWDSLGEFLALGAGFEDIGIKTGNERAKLLGKTLDAAIGKLLDN
MMAR_0158|M.marinum_M VEENHLRWDSLGEFLALGASLEDMGIKIGNDRAKILARTLDDAIGKLLGN
MUL_4939|M.ulcerans_Agy99 VEENHLRWDSLGEFLALGASLEDMGTKIGNDRAKILARTLDDAIGKLLGN
MLBr_02672|M.leprae_Br4923 VEENHLRWDSLGEFLALAVSLEDLGIKTGNERAKILAKTLDVATGKLLDN
MAV_5105|M.avium_104 VEENHLRWDSLGEYLALGACFEDTGIKTGNKRATILAKSLDAATGKLLDN
:************:***.. :** . * ** :* :*. :** * **** :
MSMEG_1654|M.smegmatis_MC2_155 NKSPSRRTGELDNRGSQFYLSLFWAQALAEQTEDAELAERFKPLAKALAE
TH_1971|M.thermoresistible__bu NKSPSRKTGELDNRGSQFYLAMYWAQALAEQDDDAELAERFAALARALEE
Mflv_3455|M.gilvum_PYR-GCK NKSPSRKAGELDNRGSQFYLAMYWAQELAEQTDDKELAEHFAPLAKTLAE
Mvan_3212|M.vanbaalenii_PYR-1 DKGPSRRTGELDNRGSQFYLAMYWAQELAEQTDDKALAEHFAPLAKTLAE
MAB_3686c|M.abscessus_ATCC_199 DKSPSRKAGELDNRGSQFYLALYWAAELAAQTEDVELAELFAPLAKQLVE
Mb0067c|M.bovis_AF2122/97 DKSPSRKTGELDNRGSQFYLAMYWAQELAAQTDDQQLAEHFASLADVLTK
Rv0066c|M.tuberculosis_H37Rv DKSPSRKTGELDNRGSQFYLAMYWAQELAAQTDDQQLAEHFASLADVLTK
MMAR_0158|M.marinum_M NKSPSRKTGELDNRGSQFYLALYWAQELAEQTDDAELAEHFAKLAKALAD
MUL_4939|M.ulcerans_Agy99 NKSPSRKTGELDNRGSQFYLALYWAQELAEQTDDAELAEHFAKLAKALAD
MLBr_02672|M.leprae_Br4923 RKSPSRKAGELDNRGSQFYLAMYWAQELATQTDDQHLRNYFTPLAETLAK
MAV_5105|M.avium_104 NKSPSRKTGELDNRGSQFYLALYWAQELAAQHDDEELRKHFAALADELRN
*.***::************:::** ** * :* * : * ** * .
MSMEG_1654|M.smegmatis_MC2_155 QEEAIVSELNSVQGKTVDIGGYYYPDPEKTSEVMRPSKTFNTTLESV---
TH_1971|M.thermoresistible__bu NEDTIVNELNEVQGKPVDIGGYYYPDPEKTAAVMRPSKTFNEVLAAAAN-
Mflv_3455|M.gilvum_PYR-GCK NEETIVEEFNKVQGESVDIGGYYYPDPEKTTAVMRPSSTFNEALESARS-
Mvan_3212|M.vanbaalenii_PYR-1 NESTIVDELNAAQGKPVDIGGYYYPDQEKTTAVMRPSATFNSALEAAGS-
MAB_3686c|M.abscessus_ATCC_199 NEDQIVAELAAVQGSPADIGGYYAPDPEKTNAVMRPSQTFNAALASVEV-
Mb0067c|M.bovis_AF2122/97 NEDVIVRELTEVQGEPVDIGGYYAPDSDMTTAVMRPSKTFNAALEAVQG-
Rv0066c|M.tuberculosis_H37Rv NEDVIVRELTEVQGEPVDIGGYYAPDSDMTTAVMRPSKTFNAALEAVQG-
MMAR_0158|M.marinum_M GEEAIVAELAEAQGEAVDIGGYYAPDNEKTTAVMRPSKTLNAALEAAQG-
MUL_4939|M.ulcerans_Agy99 GEEAIVAELAEAQGEAVDIGGYYAPDNEKTTAVMRPSKTLNAALEAAQG-
MLBr_02672|M.leprae_Br4923 NECTIVAELNEAQGKAVDIGGYYAPDAELAKAVMRPSAVFNAALAAAQGS
MAV_5105|M.avium_104 HEDTILAELAKAQGESVDIGGYYRPDDEKLTAVMRPSKTLNDALVTHRAD
* *: *: .**...****** ** : ***** .:* .* :
MSMEG_1654|M.smegmatis_MC2_155 --------
TH_1971|M.thermoresistible__bu --------
Mflv_3455|M.gilvum_PYR-GCK --------
Mvan_3212|M.vanbaalenii_PYR-1 --------
MAB_3686c|M.abscessus_ATCC_199 --------
Mb0067c|M.bovis_AF2122/97 --------
Rv0066c|M.tuberculosis_H37Rv --------
MMAR_0158|M.marinum_M --------
MUL_4939|M.ulcerans_Agy99 --------
MLBr_02672|M.leprae_Br4923 --------
MAV_5105|M.avium_104 AGRAKLVQ