For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNKFTTFPALLAVVALTALATGCGHRSSGAGQTTGDPAAAEFASGLRNKVTTDAMMTHLTKLQDIANANN GTRAVGTPGYEASVDYVVKTLRDSGFDVQTPEFSARVFHAEKPDLTVGGRPVEAHALDFSLGTGPDGVSG PLVAAPPNNPGCAVTDYGSLPVQGAVILLDRGTCPFAQKEDVAAQRGAVAMIIADNVDEQQMGGTLGPST EVKIPVLSVTKSVGVQLRGQPGPTTIKLTASAQSFKARNVIAQTKTGSEHDVVMAGAHLDSVPEGPGIND NGSGVAAVLETAVRLGSSPPVHNAVRFGFWGAEELGLIGSRNYVESLDLTALKNIALYLNFDMLASPNPG YFTYDGDQSLPMDARGQPVVPEGSAGIERTLVAYLKSAGKTAQDTSFDGRSDYDGFTLAGIPAGGLFSGA EGKMSADQAKLWGGTADQPFDPNYHQKTDTLDHIDRTALGINGGGVAYAIGLYAQDLGGHNGVPAMADRT RHVLAKS*
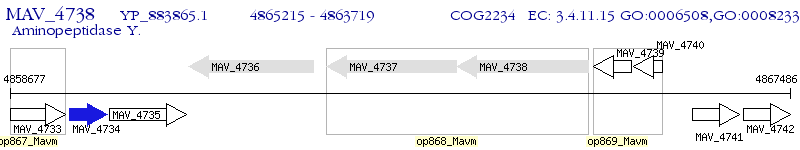
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4738 | - | - | 100% (498) | peptidase, M28 family protein |
| M. avium 104 | MAV_4737 | - | e-117 | 49.90% (483) | peptidase, M28 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0426 | lpqL | 0.0 | 80.56% (499) | lipoprotein aminopeptidase LpqL |
| M. gilvum PYR-GCK | Mflv_0190 | - | 1e-174 | 63.60% (489) | aminopeptidase Y |
| M. tuberculosis H37Rv | Rv0418 | lpqL | 0.0 | 80.56% (499) | lipoprotein aminopeptidase LpqL |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4213c | - | 1e-164 | 61.30% (491) | lipoprotein aminopeptidase LpqL |
| M. marinum M | MMAR_0726 | lpqL | 0.0 | 78.13% (503) | lipoprotein aminopeptidase LpqL |
| M. smegmatis MC2 155 | MSMEG_0806 | - | 1e-178 | 64.98% (494) | hydrolase |
| M. thermoresistible (build 8) | TH_2315 | lpqL | 1e-170 | 60.20% (495) | PROBABLE LIPOPROTEIN AMINOPEPTIDASE LPQL |
| M. ulcerans Agy99 | MUL_3587 | lpqL | 0.0 | 77.14% (503) | lipoprotein aminopeptidase LpqL |
| M. vanbaalenii PYR-1 | Mvan_0715 | - | 1e-179 | 65.05% (495) | aminopeptidase Y |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0426|M.bovis_AF2122/97 --------MVNKSRMMPAVLAVAVVVAFLTTGCIRWS--TQSR----PVV
Rv0418|M.tuberculosis_H37Rv --------MVNKSRMMPAVLAVAVVVAFLTTGCIRWS--TQSR----PVV
MMAR_0726|M.marinum_M --------MVKKSRTMPAMVAVA-VVAFLVVGCVRWS--TQTQTQPQPAT
MUL_3587|M.ulcerans_Agy99 --------MVKKSRTMPAMVAVA-VVAFLVVGCVRWS--TQTQTQPQPAI
MAV_4738|M.avium_104 --------MVNKFTTFPALLAVV-ALTALATGCGHRS--SGAG----QTT
Mflv_0190|M.gilvum_PYR-GCK MTVAGRANVVAVTRRRVAAVLAVAVLA--VAGCGRE---TAPAPQPQPD-
Mvan_0715|M.vanbaalenii_PYR-1 --------MVAVLRTRLVALLAVALLV--VAGCSRN---ADPAPQPVRD-
MSMEG_0806|M.smegmatis_MC2_155 --------MGAMRRSLTGAVLGTVVLAGALAGCDAKTELQAPQAQPTPEG
TH_2315|M.thermoresistible__bu --------LTAGALMTAMAVFGTAVVG----GCSRS-----TQTEPVPG-
MAB_4213c|M.abscessus_ATCC_199 --------MATNRVLYAVAVSAACALAT-VTACDRDA--AEAPPRSAPQA
: : . : .* .
Mb0426|M.bovis_AF2122/97 NGPAAAEFAVALRNRVSTDAMMAHLSKLQDIANANDGTRAVGTPGYQASV
Rv0418|M.tuberculosis_H37Rv NGPAAAEFAVALRNRVSTDAMMAHLSKLQDIANANDGTRAVGTPGYQASV
MMAR_0726|M.marinum_M SNPAAAEFAEALRNRVTTEAMMGHLTKLQEIADANNGTRAVGTPGYEASV
MUL_3587|M.ulcerans_Agy99 SNPAAAEFAEALRNRVTTEAMMGHLTKLQEIADANNGTRAVGTPGYEASV
MAV_4738|M.avium_104 GDPAAAEFASGLRNKVTTDAMMTHLTKLQDIANANNGTRAVGTPGYEASV
Mflv_0190|M.gilvum_PYR-GCK -PTPAGEFADELSGKVTADAMMTHLTRLQEIADAHGGNRALGSPGYRESV
Mvan_0715|M.vanbaalenii_PYR-1 -QAAAAEFADALSKKVTADAMMAHLTRLQEIADDNGGNRALGEPGYDASV
MSMEG_0806|M.smegmatis_MC2_155 VSAAAVEFANQLRERVTIDAMMGHLEKLQEIADNNDGNRALGTPGYDASV
TH_2315|M.thermoresistible__bu AAPAAADFAAELRDRISGDAVFAHLTELQRIADEHGGNRALGTPGYDASV
MAB_4213c|M.abscessus_ATCC_199 GSEEAVGFAHSLHEKVTVDNVVKHLSALQEIADKNNNTRAAGTAGFDQSV
* ** * ::: : :. ** ** **: :...** * .*: **
Mb0426|M.bovis_AF2122/97 DYVVNTLRNSGFDVQTPEFSARVFKAEKGVVTLGGNTVEARALEYSLGTP
Rv0418|M.tuberculosis_H37Rv DYVVNTLRNSGFDVQTPEFSARVFKAEKGVVTLGGNTVEARALEYSLGTP
MMAR_0726|M.marinum_M DYVVNILRNSGFDVQTPEFSARVFHSEKGSVDVGGMTAEAHALEYSLGTP
MUL_3587|M.ulcerans_Agy99 DYVVNILRNSGFDVQTPEFSARVFHSEKGSVDVGGMTAEAHALEYSLGTP
MAV_4738|M.avium_104 DYVVKTLRDSGFDVQTPEFSARVFHAEKPDLTVGGRPVEAHALDFSLGTG
Mflv_0190|M.gilvum_PYR-GCK DYVADILRDKGFDVETQDFVVKIPWADEPSLTVAGNKVEARPMMYTIGTD
Mvan_0715|M.vanbaalenii_PYR-1 GYVVDALRDKGFEVQTPTFDVRIPWADEPSLTVAGAKTAARPMEFTIGAG
MSMEG_0806|M.smegmatis_MC2_155 DYVVSALRDKGFDVETPEFEVRLPYAEDPQVTVRGQAVTAKPLEFTIGT-
TH_2315|M.thermoresistible__bu DYVVDVLRDSGFEVETPEFEVRLPFAEDPEVTVGGAPVSAKPLLFTIGTG
MAB_4213c|M.abscessus_ATCC_199 DYVVKALKDKGFDVQTPEFSFKYFQAKSLDLTVGPKKVDAGVLSYSPGG-
.**.. *::.**:*:* * : :.. : : . * : :: *
Mb0426|M.bovis_AF2122/97 PDGVTGPLVAAPADD-SPGCSPSDYDRLPVSGAVVLVDRGVCPFAQKEDA
Rv0418|M.tuberculosis_H37Rv PDGVTGPLVAAPADD-SPGCSPSDYDRLPVSGAVVLVDRGVCPFAQKEDA
MMAR_0726|M.marinum_M PDGVTGPLLLAPSDD-SPGCTPSDYDNLPVKGAVVLVDRGECQFAQKEDA
MUL_3587|M.ulcerans_Agy99 PDGVTGPLLLAPSDD-SPGCTPSDYDNLPVKGAVVLVDRGECQFAQKEDA
MAV_4738|M.avium_104 PDGVSGPLVAAPPNN--PGCAVTDYGSLPVQGAVILLDRGTCPFAQKEDV
Mflv_0190|M.gilvum_PYR-GCK GDGVSGPIVVAKSED-TPGCTADDYDGLPVDGAIVLVDRGSCPFGGKQAI
Mvan_0715|M.vanbaalenii_PYR-1 PEGVSGPLVAARSED-TPGCTADDYNGLPVQGAVVLVDRGACPFGGKQAV
MSMEG_0806|M.smegmatis_MC2_155 -EGVSGPLVPAKVED-TPGCEAGDYDGLNVAGAVVLVDRGACAFSIKQAV
TH_2315|M.thermoresistible__bu PDGVSGPLVPARADDDSPGCTADDYDGLPVDGAVVLVDRGHCPFGDKQAA
MAB_4213c|M.abscessus_ATCC_199 --RVEGRLVPARAEE-SPGCTVEDYDGLDVKGAVVLVDRGSCPFADKERV
* * :: * :: *** **. * * **::*:*** * *. *:
Mb0426|M.bovis_AF2122/97 AAQRGAVALIIADNIDEQAMG-GTLGANTDVKIPVVSVTKSVGFQLRGQS
Rv0418|M.tuberculosis_H37Rv AAQRGAVALIIADNIDEQAMG-GTLGANTDVKIPVVSVTKSVGFQLRGQS
MMAR_0726|M.marinum_M AAQRGAVALIIADNVDEQMMG-GTLGVNTDVKIPVVSVTKSVGLQLRGKS
MUL_3587|M.ulcerans_Agy99 AAQRGADALIIADNVDEQMMG-GTLGVNTDVKIPVVSVTKSVGLQLRGKS
MAV_4738|M.avium_104 AAQRGAVAMIIADNVDEQQMG-GTLGPSTEVKIPVLSVTKSVGVQLRGQP
Mflv_0190|M.gilvum_PYR-GCK AAERGAVALIVANNEDGDEFGGGTLGGDTDVKIPVVGVTREAGEQLRREP
Mvan_0715|M.vanbaalenii_PYR-1 AAERGAVALIVADSQDGGDLGAGTLGESTDVKIPVVYVTKDVGERLRGQQ
MSMEG_0806|M.smegmatis_MC2_155 AAERGAVAMIVANNTDGDEMG-GTLGRRTDVKIPVVSVTKAEGARLRANP
TH_2315|M.thermoresistible__bu AAERGAVALIVANTVDEEEMG-GTLGERTEVEIPVVSVSKSEGARLRERP
MAB_4213c|M.abscessus_ATCC_199 AAERGAVAVIIADNVDENKTS-GTLGEDSNPKIPVVSVTKSVGADLRAHP
**:*** *:*:*:. * . **** :: :***: *:: * ** .
Mb0426|M.bovis_AF2122/97 G-PTTVKLTASTQSFKARNVIAQTKTGSSANVVMAGAHLDSVPEGPGIND
Rv0418|M.tuberculosis_H37Rv G-PTTVKLTASTQSFKARNVIAQTKTGSSANVVMAGAHLDSVPEGPGIND
MMAR_0726|M.marinum_M G-PATIKLTASSQSFKARNVIAQTKTGSTSDVVMAGAHLDSVPQGPGIND
MUL_3587|M.ulcerans_Agy99 G-PATIKLTASSQSFKARNVIAQTKTGSTSDVVMAGAHLDSVPQGPGIND
MAV_4738|M.avium_104 G-PTTIKLTASAQSFKARNVIAQTKTGSEHDVVMAGAHLDSVPEGPGIND
Mflv_0190|M.gilvum_PYR-GCK GAPAVLNLNAGVRVERTQNVIAQTTTGSTADVVMLGAHLDSVSEGPGIND
Mvan_0715|M.vanbaalenii_PYR-1 GAPAVLNLNAGVRVEQSRNVIAQTKTGSTADVVMVGAHLDSVAEGPGIND
MSMEG_0806|M.smegmatis_MC2_155 G-PTTISLVAGVRVEKTRNVIAQTDTGSSSDVVMVGAHLDSVAEGPGIND
TH_2315|M.thermoresistible__bu G-PTAITLEAGVETKRTRNVIAQTSTGATSDVVMIGAHLDGVPDGPGMND
MAB_4213c|M.abscessus_ATCC_199 D-KVVLNVDAETKDVKARNVIAQTKTGATTDVVMAGAHLDSVPEGPGIND
. ..:.: * . :::****** **: :*** *****.*.:***:**
Mb0426|M.bovis_AF2122/97 NGSGVAAVLETAVQLGNSPHVSNAVRFAFWGAEEFGLIGSRNYVESLDID
Rv0418|M.tuberculosis_H37Rv NGSGVAAVLETAVQLGNSPHVSNAVRFAFWGAEEFGLIGSRNYVESLDID
MMAR_0726|M.marinum_M NGSGVAAVLETAVQLGNSPHVHNAVRVAFWGAEELGLIGSRNYVQSLDLD
MUL_3587|M.ulcerans_Agy99 NGSGVAAVLETAVQLGNSPHVHNAVRVAFWGAEELGLIGSRNYVQSLDLD
MAV_4738|M.avium_104 NGSGVAAVLETAVRLGSSPPVHNAVRFGFWGAEELGLIGSRNYVESLDLT
Mflv_0190|M.gilvum_PYR-GCK NGSGVAAILETAVQLGGAPEVANAVRFAFWGAEEDGLRGSNNYVGTLDTE
Mvan_0715|M.vanbaalenii_PYR-1 NGSGVAAVLETALQLGSTPEVANAVRFGFWGAEEEGLLGSRDYVTSLDAE
MSMEG_0806|M.smegmatis_MC2_155 NGSGVAAVLETALQLGSSPDVNYAVRFGFWGAEEEGLLGSTNYVESLNED
TH_2315|M.thermoresistible__bu NGSGVAAVLETARQLGSAPDIANAVRFGFWGAEEVGLRGSRAYLASLDVE
MAB_4213c|M.abscessus_ATCC_199 NGTGTAAVLETALQLGPSPDVKNAVRFAFWGAEEEGLIGSTDYVKSLDVA
**:*.**:**** :** :* : ***..****** ** ** *: :*:
Mb0426|M.bovis_AF2122/97 ALKGIALYLNFDMLASPNPGYFTYDGDQSLPLDAR-GQPVVPEGSAGIER
Rv0418|M.tuberculosis_H37Rv ALKGIALYLNFDMLASPNPGYFTYDGDQSLPLDAR-GQPVVPEGSAGIER
MMAR_0726|M.marinum_M ELKNIALYLNFDMLASPNPGYFTYDGDQSLPLDER-GQPVVPEGSAGIER
MUL_3587|M.ulcerans_Agy99 ELKNIALYLNFDMLASPNPGIFTYDGDQSLPLVEH-GQPVVPEGSAGIER
MAV_4738|M.avium_104 ALKNIALYLNFDMLASPNPGYFTYDGDQSLPMDAR-GQPVVPEGSAGIER
Mflv_0190|M.gilvum_PYR-GCK TLKDIALYLNFDMLGSPNPGYFTYDGNQSTAPDPRIGVPRVPEGSAGIER
Mvan_0715|M.vanbaalenii_PYR-1 ALKDIALYLNFDMLGSPNPGYFTMDGNQSTPPGQGEGVPRVPEGSAGIER
MSMEG_0806|M.smegmatis_MC2_155 QLKDIALYLNFDMLGSPNPGYFTYDGDQSAPPEPR--VPRVPEGSAGIER
TH_2315|M.thermoresistible__bu ELKDIAVYLNLDMVGSPNPGYFTYDGDLSSPPDPNRPAPRVPEGSAGVER
MAB_4213c|M.abscessus_ATCC_199 ALKNIALYLNYDMLGSPNAAYLTYDGDQSNEPDPNEIPVRIPEGSAGIER
**.**:*** **:.***.. :* **: * :******:**
Mb0426|M.bovis_AF2122/97 TFVAYLKMAGKTAQDTSFDGRSDYDGFTLAGIPSGGLFSGAEVKKSAEQA
Rv0418|M.tuberculosis_H37Rv TFVAYLKMAGKTAQDTSFDGRSDYDGFTLAGIPSGGLFSGAEVKKSAEQA
MMAR_0726|M.marinum_M TLVSYLRLSGKIAQDTSFDGRSDYDGFTLAGIPSGGLFSGAEVKMTEEQA
MUL_3587|M.ulcerans_Agy99 TLVSYLRLSGKIAQDTSFDGRSDYDGFTLAGIPSGGLFSGAEVKMTEEQA
MAV_4738|M.avium_104 TLVAYLKSAGKTAQDTSFDGRSDYDGFTLAGIPAGGLFSGAEGKMSADQA
Mflv_0190|M.gilvum_PYR-GCK TLAAYLDRAGKPARDTSFDGRSDYDAFTKAGVPAGGLFSGAEAKMSSEEA
Mvan_0715|M.vanbaalenii_PYR-1 MLMAYLDRAGKPAQDTSFNGRSDYDGFTMVGVPAGGLFSGAEDKMSAEQA
MSMEG_0806|M.smegmatis_MC2_155 TLAAYLADAGKMPQDTSFDGRSDYDAFTLAGIPAGGLFSGAEEKMSPEEA
TH_2315|M.thermoresistible__bu TLVAALADEDIEPDDVNLEGRSDNDSFTLAGIPAGGLFTGGSGEKTEEQA
MAB_4213c|M.abscessus_ATCC_199 TEVAYLAEQGKKAHDTGYDGRSDYDAFSRAGIPTGGIFSGAEDKMSDEEA
: * . . *.. :**** *.*: .*:*:**:*:*.. : : ::*
Mb0426|M.bovis_AF2122/97 ELWGGTADEPFDPNYHQKTDTLDHIDRTALGINGAGVAYAVGLYAQDLGG
Rv0418|M.tuberculosis_H37Rv ELWGGTADEPFDPNYHQKTDTLDHIDRTALGINGAGVAYAVGLYAQDLGG
MMAR_0726|M.marinum_M KLWGGTADEPFDPNYHQKSDTIDHIDRTSLGIQGGGVAYAVGLYAQDLTG
MUL_3587|M.ulcerans_Agy99 KLWGGTADEPFDPNYHQKSDTIDHIDRTSLGIQGGGVAYAVGLYAQDLTG
MAV_4738|M.avium_104 KLWGGTADQPFDPNYHQKTDTLDHIDRTALGINGGGVAYAIGLYAQDLGG
Mflv_0190|M.gilvum_PYR-GCK ELWGGEADQPFDPNYHKASDTLEHIDRTALQIHGSGVAYAAGLYAQDQRG
Mvan_0715|M.vanbaalenii_PYR-1 EIWGGRADEPFDPNYHKASDTLEHIDRTALQVHGSGVAYSVGLYAQDQRG
MSMEG_0806|M.smegmatis_MC2_155 EEWGGAADEPFDPNYHKNTDTLEHVDRDALQIHGEGVAFTVGLYSQDQRG
TH_2315|M.thermoresistible__bu ERWGGTAGEPFDPNYHKEGDDLDNIDRTALAVNAAAAAYAVGMYAQDQRG
MAB_4213c|M.abscessus_ATCC_199 KQWGGKAGQPFDPNYHQAGDTLANVNKDALKINAGGVAYTVGLYAQSIDG
: *** *.:*******: * : :::: :* ::. ..*:: *:*:*. *
Mb0426|M.bovis_AF2122/97 PNGVPVMADRTRHLIAKP
Rv0418|M.tuberculosis_H37Rv PNGVPVMADRTRHLIAKP
MMAR_0726|M.marinum_M RNGVPVIGDRTRHVLAKP
MUL_3587|M.ulcerans_Agy99 RNGVPVIGDRTRHVLAKP
MAV_4738|M.avium_104 HNGVPAMADRTRHVLAKS
Mflv_0190|M.gilvum_PYR-GCK RNGIPVREDRTRHQLTGP
Mvan_0715|M.vanbaalenii_PYR-1 RNGIPIREDRIRHKLTAP
MSMEG_0806|M.smegmatis_MC2_155 RNGIPVRDDRTRHQISPQ
TH_2315|M.thermoresistible__bu RNGLPIRDDRTRHQLSNP
MAB_4213c|M.abscessus_ATCC_199 RNGVPVHEDRTRHQLKG-
**:* ** ** :