For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IDHVGINCANYAESQRFYDAVLGTLGFSRQLDVGPAIGYGRAGKPTFWISDAPAGPNREVHLAFEAADED AVRAFHRAAVELGAEVLHPPRLWPEYHPGYFGAFVRDPDGNNVEAVWHGA*
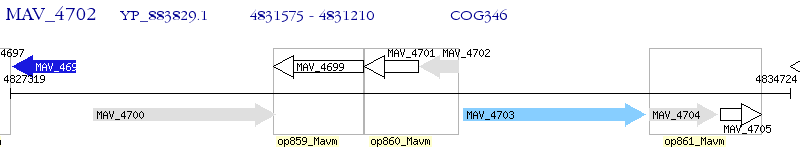
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4702 | - | - | 100% (121) | glyoxalase family protein |
| M. avium 104 | MAV_0150 | - | 3e-14 | 32.03% (128) | glyoxalase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0135 | - | 6e-46 | 66.13% (124) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0642 | - | 4e-42 | 63.20% (125) | putative glyoxalase/bleomycin resistance protein |
| M. marinum M | MMAR_3611 | - | 1e-07 | 30.89% (123) | hypothetical protein MMAR_3611 |
| M. smegmatis MC2 155 | MSMEG_0884 | - | 1e-48 | 69.60% (125) | glyoxalase family protein |
| M. thermoresistible (build 8) | TH_0973 | - | 2e-50 | 71.20% (125) | glyoxalase family protein |
| M. ulcerans Agy99 | MUL_1255 | - | 1e-06 | 30.83% (120) | hypothetical protein MUL_1255 |
| M. vanbaalenii PYR-1 | Mvan_0768 | - | 5e-47 | 66.40% (125) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0884|M.smegmatis_MC2_155 -----MIDHFGINCVDFEKSKTFYDKVLGVLG----YTRQMDFDGAIGYG
Mvan_0768|M.vanbaalenii_PYR-1 -----MIDHFGINCADWETSKAFYDKVLGVLG----YTRQMDFQVAVGYG
Mflv_0135|M.gilvum_PYR-GCK MYGGTVIDHFGINCADWERSKAFYDKVLGVLG----YTRQMDFGVAIGYG
TH_0973|M.thermoresistible__bu -----VIDHLGINCADFAKSTEFYDKVLGVLG----YTRQLDYEVAIGYG
MAV_4702|M.avium_104 -----MIDHVGINCANYAESQRFYDAVLGTLG----FSRQLDVGPAIGYG
MAB_0642|M.abscessus_ATCC_1997 -----MIDHFGINCADLPAAAAFYDKVLGVLG----FTRQMDFGVAIGYG
MMAR_3611|M.marinum_M -MRWRGVHHVEFNVLDYDKSIAFYDAMFGWLGYMSFWTLDIEYRSTYYMA
MUL_1255|M.ulcerans_Agy99 ---------MEFNVLDYDKSIAFYDAMFGWLGYMSFWTLDIEYRSTYYMA
. :* : : *** ::* ** :: ::: : .
MSMEG_0884|M.smegmatis_MC2_155 ADGHPDFWIA-DAGAGNVAGPNRE-----VHIAFKASGVDAVQAFYDAAL
Mvan_0768|M.vanbaalenii_PYR-1 VEGKPDFWIA-DMSSGDAAGPNRE-----VHIAFQAAGVEAVQAFYDAAL
Mflv_0135|M.gilvum_PYR-GCK TDGHPDFWIA-DMSTGAAAGPNRE-----VHIAFAAKDAESVQAFFRTAL
TH_0973|M.thermoresistible__bu RDGRPSFWIA-DASAGDVTGPNRE-----VHVAFRADDEASVQAFYRTAL
MAV_4702|M.avium_104 RAGKPTFWIS-DAP----AGPNRE-----VHLAFEAADEDAVRAFHRAAV
MAB_0642|M.abscessus_ATCC_1997 PEGQAQFWIGGDPSGHASMESNRE-----VHVAFSAASTAKVDEFFAAAT
MMAR_3611|M.marinum_M RFPLPHSYIGIQPADGGGQLRHRDQKVGINHIALWARSRAEIDRFHRDFL
MUL_1255|M.ulcerans_Agy99 RFPLPHSYIGIQPADGGGQLRHRDQKVGINHIALWARSRAEIDRFHRDFL
. :*. : :*: *:*: * . : *.
MSMEG_0884|M.smegmatis_MC2_155 ELGAESLHAP-RLWPEYHPGYFGAFVRDPDG-------------------
Mvan_0768|M.vanbaalenii_PYR-1 DAGAESLHAP-RLWPEYHPGYYGAFVRDPDG-------------------
Mflv_0135|M.gilvum_PYR-GCK ALGVEPLHEP-RLWPEYHENYYGAFVRDPDG-------------------
TH_0973|M.thermoresistible__bu ALGAEPLHEP-RLWPEYHPGYYGAFVRDPDG-------------------
MAV_4702|M.avium_104 ELGAEVLHPP-RLWPEYHPGYFGAFVRDPDG-------------------
MAB_0642|M.abscessus_ATCC_1997 GAGAEVLHAP-RLWPEYHPGYYGAFVRDPDG-------------------
MMAR_3611|M.marinum_M LPRAIPVTDPPREYPIYTPGYYAVFFDDPINGIHWELARLPSVPSPRDFW
MUL_1255|M.ulcerans_Agy99 LPRAIPVTDPPREYPIYTPGYYAVFFDDPINGIHWELARLPSVPSPRDFW
. : * * :* * .*:..*. ** .
MSMEG_0884|M.smegmatis_MC2_155 ---NNVEAVFHGAQ--------------------
Mvan_0768|M.vanbaalenii_PYR-1 ---NNVEAVFHGA---------------------
Mflv_0135|M.gilvum_PYR-GCK ---NNVEAVFHGGGPSGGPPGPRPA---------
TH_0973|M.thermoresistible__bu ---NNVEAVFHGATPQD-----------------
MAV_4702|M.avium_104 ---NNVEAVWHGA---------------------
MAB_0642|M.abscessus_ATCC_1997 ---NNVEAVFHG----------------------
MMAR_3611|M.marinum_M KSYRALRAVSADHPEWKRSVAREAMRSLPGRTSR
MUL_1255|M.ulcerans_Agy99 KSYRTLRAVSADHPEWKRSVAREAMRSLPGRTSR
. :.** .