For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAGGEPVTVGALTRADARRCAELEAQLFDGDDPWPAAAFHRELASAHNHYVGARVGDTLVGYAGISRLGR VPPYEYEIHTIGVDPAYQGRGIGRLMLDRLLEFADGGVVYLEVRTDNEPAIGLYRSVGFEQIGLRRRYYR ISGADAYTMRRKARQ*
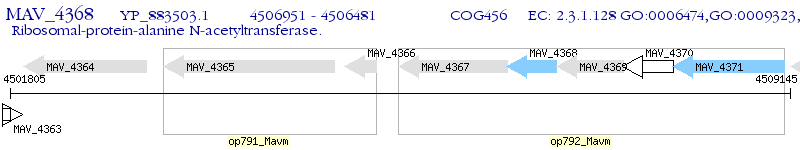
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4368 | rimI | - | 100% (156) | ribosomal-protein-alanine acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3454c | rimI | 3e-71 | 81.58% (152) | ribosomal-protein-alanine acetyltransferase |
| M. gilvum PYR-GCK | Mflv_4930 | - | 3e-59 | 71.92% (146) | ribosomal-protein-alanine acetyltransferase |
| M. tuberculosis H37Rv | Rv3420c | rimI | 3e-71 | 81.58% (152) | ribosomal-protein-alanine acetyltransferase |
| M. leprae Br4923 | MLBr_00378 | - | 1e-62 | 69.68% (155) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_3735c | - | 2e-49 | 68.06% (144) | ribosomal-protein-alanine acetyltransferase RimI |
| M. marinum M | MMAR_1123 | rimI | 3e-73 | 84.87% (152) | ribosomal-protein-alanine acetyltransferase, RimI |
| M. smegmatis MC2 155 | MSMEG_1579 | rimI | 2e-56 | 72.86% (140) | ribosomal-protein-alanine acetyltransferase |
| M. thermoresistible (build 8) | TH_1690 | rimI | 1e-59 | 73.33% (150) | PROBABLE RIBOSOMAL-PROTEIN-ALANINE ACETYLTRANSFERASE RIMI |
| M. ulcerans Agy99 | MUL_0883 | rimI | 2e-73 | 84.87% (152) | ribosomal-protein-alanine acetyltransferase, RimI |
| M. vanbaalenii PYR-1 | Mvan_1488 | - | 4e-58 | 70.27% (148) | ribosomal-protein-alanine acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3454c|M.bovis_AF2122/97 --------------------------------------------------
Rv3420c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1123|M.marinum_M --------------------------------------------------
MUL_0883|M.ulcerans_Agy99 --------------------------------------------------
MAV_4368|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 MSIVLAIDTATAAVTAGIVAFDGHDCFTLAERVTVDAKAHVERLTPNVLV
TH_1690|M.thermoresistible__bu --------------------------------------------------
Mflv_4930|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1488|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1579|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3735c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3454c|M.bovis_AF2122/97 --------------------------------------------------
Rv3420c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1123|M.marinum_M --------------------------------------------------
MUL_0883|M.ulcerans_Agy99 --------------------------------------------------
MAV_4368|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 ALADAELAMCELDAVVVGCGPGPFTGLRVGMATAAAYGHALGIPVHGVCS
TH_1690|M.thermoresistible__bu --------------------------------------------------
Mflv_4930|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1488|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1579|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3735c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3454c|M.bovis_AF2122/97 --------------------------------------------------
Rv3420c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1123|M.marinum_M --------------------------------------------------
MUL_0883|M.ulcerans_Agy99 --------------------------------------------------
MAV_4368|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 LDAIGVRTTGDTLVVTDARRHEVYWARYRDGVRIAGPAVGSPTDVDPGTA
TH_1690|M.thermoresistible__bu --------------------------------------------------
Mflv_4930|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1488|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1579|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3735c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3454c|M.bovis_AF2122/97 --------------------------------------------------
Rv3420c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1123|M.marinum_M --------------------------------------------------
MUL_0883|M.ulcerans_Agy99 --------------------------------------------------
MAV_4368|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 LTVAGSPEHAALFGLPLCEPIYPTPAGLVAAVPDWSVSPIPLVALYLRRP
TH_1690|M.thermoresistible__bu --------------------------------------------------
Mflv_4930|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1488|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1579|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3735c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3454c|M.bovis_AF2122/97 ----MTADTEP----VTIGALTRADAQRCAELEAQLFVGDDPWPPAAFNR
Rv3420c|M.tuberculosis_H37Rv ----MTADTEP----VTIGALTRADAQRCAELEAQLFVGDDPWPPAAFNR
MMAR_1123|M.marinum_M ----MTADLGP----VTIGALTRADARRCAELEAQLFDGDDPWPEVAFER
MUL_0883|M.ulcerans_Agy99 ----MTADLGP----VTIGALTRADARRCAELEAQLFDGDDPWPEVAFER
MAV_4368|M.avium_104 ----MTAGGEP----VTVGALTRADARRCAELEAQLFDGDDPWPAAAFHR
MLBr_00378|M.leprae_Br4923 DAKPITADNEP----IVIGTLTPADVDRCAQLESQFFDGDNPWPAAAFDR
TH_1690|M.thermoresistible__bu ----MTAPDRDRDTGVVLTPLTPADARRCAELELELFGGDDPWPVAAFRA
Mflv_4930|M.gilvum_PYR-GCK ----MSAEYGP---------LQTTDALRCAELEAQLFDGDDPWPERAFLA
Mvan_1488|M.vanbaalenii_PYR-1 ----MNVEYGP---------LTGADAARCAELEAQLFDGDDPWPERAFLA
MSMEG_1579|M.smegmatis_MC2_155 MSIDGTVQYTP---------LHKDDAARCAELESQLFGGDDPWPARAFLA
MAB_3735c|M.abscessus_ATCC_199 ----MTIELKP---------LRRRDARRCAELEAVLFEGDDPWPESAFLA
. * *. ***:** :* **:*** **
Mb3454c|M.bovis_AF2122/97 ELASPHNHYVGAR-----SGGTLVGYAGISRLGRTPPFEYEVHTIGVDPA
Rv3420c|M.tuberculosis_H37Rv ELASPHNHYVGAR-----SGGTLVGYAGISRLGRTPPFEYEVHTIGVDPA
MMAR_1123|M.marinum_M ELASGRNHYVGAR-----CADNLVGYAGVSRLGRVAPFEYEIHTIGVDPA
MUL_0883|M.ulcerans_Agy99 ELASGRNHYVGAR-----CADNLVGYAGVSRLGRVAPFEYEIHTIGVDPA
MAV_4368|M.avium_104 ELASAHNHYVGAR-----VGDTLVGYAGISRLGRVPPYEYEIHTIGVDPA
MLBr_00378|M.leprae_Br4923 ELANSYNCYVGAR-----TADTLVGYAGITRLGHTPPFEYEVHTIAVDPA
TH_1690|M.thermoresistible__bu EIAVPHNRYLAARDTTRPTGDLLVGYGGISRLGRKPPYEYEIHTIAIDPA
Mflv_4930|M.gilvum_PYR-GCK ELAAKHNHYVGAR-----VDDKLVGYAGIARLGRFKPFEYEIHTIGVDPA
Mvan_1488|M.vanbaalenii_PYR-1 ELAAKHNYYVAAR-----VDDKVVGYAGIARLGRFKPYEYEIHTVGVDPA
MSMEG_1579|M.smegmatis_MC2_155 ELAAKHNHYIAAR-----CDGKLVGYAGIARLGRAVPYEYEIHTIGVDPD
MAB_3735c|M.abscessus_ATCC_199 ELASSHIYYLAAR-----DGHTLVGYGGISRLGRHAP-EYEIHTIGVDPA
*:* *:.** :***.*::***: * ***:**:.:**
Mb3454c|M.bovis_AF2122/97 YQGRGIGRRLLRELLDFARG--GVVYLEVRTDNDAALALYRSVGFQRVGL
Rv3420c|M.tuberculosis_H37Rv YQGRGIGRRLLRELLDFARG--GVVYLEVRTDNDAALALYRSVGFQRVGL
MMAR_1123|M.marinum_M YQGRGIGRRLLDELLAFADG--GVVFLEVRTDNEPAIALYRSVGFEQVGL
MUL_0883|M.ulcerans_Agy99 YQGRGIGRRLLDELLAFADG--GVVFLEVRTDNEPAIALYRSVGFEQVGL
MAV_4368|M.avium_104 YQGRGIGRLMLDRLLEFADG--GVVYLEVRTDNEPAIGLYRSVGFEQIGL
MLBr_00378|M.leprae_Br4923 YRGRGVGRRLLGELLDFAGS--GAIYLEVRTDNETAIALYRSVGFERIGL
TH_1690|M.thermoresistible__bu YQGRGIGRRLLDELLAFADG--ATVYLEVRTDNEPAINLYESAGFTRIGL
Mflv_4930|M.gilvum_PYR-GCK YQGQGIGRQMLHRLFDFADG--APIFLEVRTDNAAAIALYESEGFVKMGV
Mvan_1488|M.vanbaalenii_PYR-1 WQGQGIGRGMLSRLLDYAGD--GTVFLEVRTDNAAAIALYESEGFVRVGI
MSMEG_1579|M.smegmatis_MC2_155 HQGRGIGRAMMQLLLEYASG--GTVFLEVRTDNEPAIKLYESLGFVTIGL
MAB_3735c|M.abscessus_ATCC_199 YQKQGLGRRLLDALLAHADQDPGPVFLEVRTDNAAAIALYQGAGFETVGL
: :*:** :: *: .* . ::******* .*: **.. ** :*:
Mb3454c|M.bovis_AF2122/97 RRRYYRVSGADAYTMRRDSGDPS-
Rv3420c|M.tuberculosis_H37Rv RRRYYRVSGADAYTMRRDSGDPS-
MMAR_1123|M.marinum_M RRRYYRVSGADAYTMRRDPGGAR-
MUL_0883|M.ulcerans_Agy99 RRRYYRVSGADAYTMRRDPGGAR-
MAV_4368|M.avium_104 RRRYYRISGADAYTMRRKARQ---
MLBr_00378|M.leprae_Br4923 RPRYYPASGADAYLMRREAQ----
TH_1690|M.thermoresistible__bu RRRYYRQSGADAYTMRRDPT----
Mflv_4930|M.gilvum_PYR-GCK RKRYYRVSGADAYTMKREKRERQ-
Mvan_1488|M.vanbaalenii_PYR-1 RKRYYRVSGADAYTMKRERR----
MSMEG_1579|M.smegmatis_MC2_155 RKRYYRASGADAYTMQRLPQEGPK
MAB_3735c|M.abscessus_ATCC_199 RKRYYPGSGADAFTMKRPAHGEVK
* *** *****: *:*