For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KLPLLGPVSLAGFQHAWFFLALLAVLLVIGLYVVQQFARRRRVLRFANMEVLERVAPPHPSRWRHVPTIL LATSLVLLTTAMAGPTSDVRIPLNRAVVMLVIDVSESMASTDVPPNRLAAAKEAGKQFADQLTPAINLGL VEFAANATLLVPPTTNRAAVKAGIDSLQPAPKTATGEGIFTALQAIATVGSVMGGGEGPPPARIVLESDG AENVPLDPNAPQGAFTAARAAKAEGVQISTISFGTPYGTVDYEGATIPVPVDDQTLQKICEITDGQAFHA DSLDSLKNVYSTLQRQIGYETVKGDASMAWMLLGAVVLAGAVLAGLLLNRRLPA*
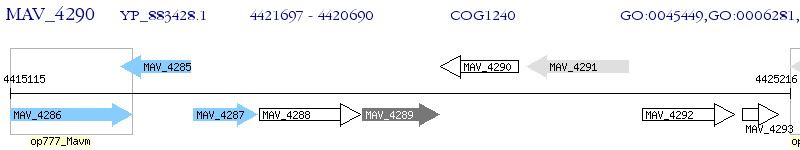
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4290 | - | - | 100% (335) | hypothetical protein MAV_4290 |
| M. avium 104 | MAV_3297 | - | e-127 | 67.07% (334) | hypothetical protein MAV_3297 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1517 | - | 1e-123 | 64.67% (334) | hypothetical protein Mb1517 |
| M. gilvum PYR-GCK | Mflv_3659 | - | 1e-123 | 65.27% (334) | hypothetical protein Mflv_3659 |
| M. tuberculosis H37Rv | Rv1481 | - | 1e-123 | 64.67% (334) | hypothetical protein Rv1481 |
| M. leprae Br4923 | MLBr_01808 | - | 1e-121 | 64.07% (334) | hypothetical protein MLBr_01808 |
| M. abscessus ATCC 19977 | MAB_2724c | - | 1e-111 | 59.64% (332) | hypothetical protein MAB_2724c |
| M. marinum M | MMAR_2288 | - | 1e-123 | 64.37% (334) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_3149 | - | 1e-121 | 65.95% (326) | hypothetical protein MSMEG_3149 |
| M. thermoresistible (build 8) | TH_1812 | - | 1e-117 | 66.56% (314) | PROBABLE MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1490 | - | 1e-124 | 65.27% (334) | hypothetical protein MUL_1490 |
| M. vanbaalenii PYR-1 | Mvan_2751 | - | 1e-125 | 66.17% (334) | hypothetical protein Mvan_2751 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1517|M.bovis_AF2122/97 -MTLPLLGPMTLSGFAHSWFFLFLFVVAGLVALYILMQLARQRRMLRFAN
Rv1481|M.tuberculosis_H37Rv -MTLPLLGPMTLSGFAHSWFFLFLFVVAGLVALYILMQLARQRRMLRFAN
MLBr_01808|M.leprae_Br4923 -MTLPLLGPMSLSGFEHSWFFLFIFIVFGLAAFYVMMQVARQRRMLRFAN
MMAR_2288|M.marinum_M -MTLPLLGPMTLSGFAHSWFFLFIFVIAGLIAVYVVLQLARQKRMLRFAN
MUL_1490|M.ulcerans_Agy99 -MTLPLLGPMTLSGFAHSWFFLFLLVVAGLIAIYVVLQLARQKRMLRFAN
Mflv_3659|M.gilvum_PYR-GCK -MTLPLLGPMSLSGFEHPWFFLFFFVVIGLVALYIVVQLARQKRMLRFAN
Mvan_2751|M.vanbaalenii_PYR-1 -MTLPLLGPMSLSGFEHPWFFLFFLVVLGLVALYVIVQMGRHRRMLRFAN
MSMEG_3149|M.smegmatis_MC2_155 ---------MSLSGFAHAWWFLFLVVVAGLVALYIVVQRARQQRILRFAN
TH_1812|M.thermoresistible__bu ---------MSLTGFEHAWFFLFFLVVLGIIALYIVVQRARHKRMLRFAN
MAB_2724c|M.abscessus_ATCC_199 MNDVPFFGPMTLTGFAHPWFFLVLLLVAALIALYVVAQYLRQKRVLRFAN
MAV_4290|M.avium_104 -MKLPLLGPVSLAGFQHAWFFLALLAVLLVIGLYVVQQFARRRRVLRFAN
::*:** *.*:** :. : : ..*:: * *::*:*****
Mb1517|M.bovis_AF2122/97 MELLESVAPKRPSRWRHVPAILLVLSLLLFTIAMAGPTHDVRIPRNRAVV
Rv1481|M.tuberculosis_H37Rv MELLESVAPKRPSRWRHVPAILLVLSLLLFTIAMAGPTHDVRIPRNRAVV
MLBr_01808|M.leprae_Br4923 MELLESVAPNRPVQWRHVPAILLMLALLLFTIAMAGPTNDVRIPRNRAVV
MMAR_2288|M.marinum_M MELLESVAPQRPSRFRHIPAMLLALSLVLFTVAMAGPTHDVRIPRNRAVV
MUL_1490|M.ulcerans_Agy99 MELLESVAPQRPSRYRHIPAMLLALSLVLFTVAMAGPTHDVRIPRNRAVV
Mflv_3659|M.gilvum_PYR-GCK MELLESVAPKQPSRMRHLPAVLMILSLVSFTIAMAGPTHDVRIPRNRAVV
Mvan_2751|M.vanbaalenii_PYR-1 MELLESVAPKRPSRWRHLPAVLLILSLMSFTVAMAGPTHDVRIPRNRAVV
MSMEG_3149|M.smegmatis_MC2_155 MELLESVAPKQPTRWRHLPAILMVCSLVLFTIAMAGPTHDVRIPRNRAVV
TH_1812|M.thermoresistible__bu MELLESVAPRRPSRWRHLPATLLALSLVVLTVAMAGPTHDIRIPRNRAVV
MAB_2724c|M.abscessus_ATCC_199 TELLDSVAPTRPRTWRHAATALLVVGLMVLTIALAGPTHDVRIPRNRAVV
MAV_4290|M.avium_104 MEVLERVAPPHPSRWRHVPTILLATSLVLLTTAMAGPTSDVRIPLNRAVV
*:*: *** :* ** .: *: .*: :* *:**** *:*** *****
Mb1517|M.bovis_AF2122/97 MLVIDVSQSMRATDVEPSRMVAAQEAAKQFADELTPGINLGLIAYAGTAT
Rv1481|M.tuberculosis_H37Rv MLVIDVSQSMRATDVEPSRMVAAQEAAKQFADELTPGINLGLIAYAGTAT
MLBr_01808|M.leprae_Br4923 MLVIDVSQSMRATDVEPNRMAAAQEAAKQFAGELTPGINLGLIAYAGTAT
MMAR_2288|M.marinum_M MLVIDVSQSMRATDVEPNRMVAAQEAAKQFADELTPGINLGLIAYAGTAT
MUL_1490|M.ulcerans_Agy99 MLVIDVSQSMRATDVEPNRMVAAQEAAKQFADELTPGINLGLIAYAGTAT
Mflv_3659|M.gilvum_PYR-GCK MLVIDVSQSMRATDVAPNRLTAAQEAAKQFADQLTPGINLGLIAYAGTAT
Mvan_2751|M.vanbaalenii_PYR-1 MLVIDVSQSMRATDVAPNRLVAAQEAAKQFADQLTPGINLGLIAYAGTAT
MSMEG_3149|M.smegmatis_MC2_155 MLVIDVSQSMRATDVAPSRLVAAQEAAKQFADQLTPGINLGLIAYAGTAT
TH_1812|M.thermoresistible__bu MLVIDVSQSMRATDVEPSRLVAAQEAAKQFADELTPGINLGLISYAGTAT
MAB_2724c|M.abscessus_ATCC_199 MLVIDVSRSMESTDVAPNRLGAAKEAGKEFARNLTPGINLGLIAYAGTAT
MAV_4290|M.avium_104 MLVIDVSESMASTDVPPNRLAAAKEAGKQFADQLTPAINLGLVEFAANAT
*******.** :*** *.*: **:**.*:** :***.*****: :*..**
Mb1517|M.bovis_AF2122/97 VLVSPTTNREATKNALDKLQFADRTATGEAIFTALQAIATVGAVIGGGDT
Rv1481|M.tuberculosis_H37Rv VLVSPTTNREATKNALDKLQFADRTATGEAIFTALQAIATVGAVIGGGDT
MLBr_01808|M.leprae_Br4923 VLVSPTTNRYATKNALDKLQFADRTATGEAIFTALQAIATVGAVIGGGEM
MMAR_2288|M.marinum_M VLVSPTTNREATKAALDKLQFADRTATGEAIFTALQAIATVGAVIGGGDT
MUL_1490|M.ulcerans_Agy99 VLVSPTTNREATKAALDKLQFADRTATGEAIFTALQAIATVGAVIGGGDT
Mflv_3659|M.gilvum_PYR-GCK VLVSPTTNRESTKTAIDKLQLADRTATGEGIFTALQAIATVGAVIGGGDE
Mvan_2751|M.vanbaalenii_PYR-1 VLVSPTTNREATKAAIDKLQLADRTATGEGIFTALQAVATVGAVIGGGDE
MSMEG_3149|M.smegmatis_MC2_155 VLVQPTTNREATKNGLDKLQLADRTATGEGIFTALQAIATVGAVIGGGDE
TH_1812|M.thermoresistible__bu VLVSPTTDREATKNAIDRLQLADRTATGEGIFTALQAIATVGAVIGGGDE
MAB_2724c|M.abscessus_ATCC_199 VLVSPTTNRDATVNALDNLQLADRTATGEGIFTALQAIATVGAVIGGGDK
MAV_4290|M.avium_104 LLVPPTTNRAAVKAGIDSLQPAPKTATGEGIFTALQAIATVGSVMGGGEG
:** ***:* :. .:* ** * :*****.*******:****:*:***:
Mb1517|M.bovis_AF2122/97 PPPARIVLFSDGKETMPTNPDNPKGAYTAARTAKDQGVPISTISFGTPYG
Rv1481|M.tuberculosis_H37Rv PPPARIVLFSDGKETMPTNPDNPKGAYTAARTAKDQGVPISTISFGTPYG
MLBr_01808|M.leprae_Br4923 PPPARIVLFSDGKETMPTNPDNPKGAYTAARTAKDQGVPISTISFGTVYG
MMAR_2288|M.marinum_M PPPARIVLFSDGKETMPTNPDNPKGAYTAARTAKDQGVPISTISFGTPYG
MUL_1490|M.ulcerans_Agy99 PPPARIVLFSDGKETMPTNPDNPKGAYTAARTAKDQGVPISTISFGTPYG
Mflv_3659|M.gilvum_PYR-GCK PPPARVVLMSDGKETVPSNPDNPKGAYTAARTAKDQGVPISTVSFGTPYG
Mvan_2751|M.vanbaalenii_PYR-1 PPPARIVLMSDGKETVPSNPDNPKGAYTAARTAKDQGVPISTVSFGTPYG
MSMEG_3149|M.smegmatis_MC2_155 PPPARIVLMSDGKETVPSNPDNPKGAFTAARTAKDQGVPISTVSFGTPYG
TH_1812|M.thermoresistible__bu PPPARIVLLSDGKETVPSNPDNPKGAYTAARTAKDQGVPISTISFGTPYG
MAB_2724c|M.abscessus_ATCC_199 PPPARIVLMSDGKETVPSNPDNPKGAYTAARTAKDQQVPISTIAFGTKDG
MAV_4290|M.avium_104 PPPARIVLESDGAENVPLDPNAPQGAFTAARAAKAEGVQISTISFGTPYG
*****:** *** *.:* :*: *:**:****:** : * ***::*** *
Mb1517|M.bovis_AF2122/97 FVEINDQRQPVPVDDETMKKVAQLSGGNSYNAATLAELRAVYSSLQQQIG
Rv1481|M.tuberculosis_H37Rv FVEINDQRQPVPVDDETMKKVAQLSGGNSYNAATLAELRAVYSSLQQQIG
MLBr_01808|M.leprae_Br4923 FVEINGQRQPVPVDDETMKKVAQLSGGNSYNAATLAELKAVYASLQQQIG
MMAR_2288|M.marinum_M FVEINDQRQPVPVDDETMKKVAQLSGGNSYNAATLAELNSVYASLQQQIG
MUL_1490|M.ulcerans_Agy99 FVEINDQRQPVPVDDETMKKVAQLSGGNSYNAATLAELNSVYVSLQQQIG
Mflv_3659|M.gilvum_PYR-GCK YVEINEQRQPVPVDDEMLKKIADLSGGEAFTASSLEQLKQVFTNLQEQIG
Mvan_2751|M.vanbaalenii_PYR-1 YVEINDQRQPVPVDDEMLKKIADLSGGDAFTASSLEQLKQVFTNLQEQIG
MSMEG_3149|M.smegmatis_MC2_155 YVEINDQRQPVPVDDEMLEKIAQLSGGDAFTASSLEQLKAVFTSLQQQIG
TH_1812|M.thermoresistible__bu YVEINEQRQPVPVDDEMLERIADLSGGSAYNAATLEQLKEVYGTLQEQIG
MAB_2724c|M.abscessus_ATCC_199 YVEINGQRQNVPYAPDMMEKVAKLSGGETYTASTLGQLKEVYANLQQQIG
MAV_4290|M.avium_104 TVDYEGATIPVPVDDQTLQKICEITDGQAFHADSLDSLKNVYSTLQRQIG
*: : ** : ::::..::.*.:: * :* .*. *: .**.***
Mb1517|M.bovis_AF2122/97 YETIKGDASVGWLRLGALALALAALAALLIN------------------R
Rv1481|M.tuberculosis_H37Rv YETIKGDASVGWLRLGALALALAALAALLIN------------------R
MLBr_01808|M.leprae_Br4923 YETIKGDASAGWLRLGVLVLALAALTALLIN------------------R
MMAR_2288|M.marinum_M YETIRGDASMGWLRLGALVLVAAALAALLIN------------------R
MUL_1490|M.ulcerans_Agy99 YETIRGDASMGWLRLGALVLVAAALAALLIN------------------R
Mflv_3659|M.gilvum_PYR-GCK YETIKGDASVGWLRLGAGVLALAALGALLIN------------------R
Mvan_2751|M.vanbaalenii_PYR-1 YETIKGDASVGWLRIGSLVLALAALGALLIN------------------R
MSMEG_3149|M.smegmatis_MC2_155 YETIKGDASVGWLRLGALILALAGVAALLIN------------------R
TH_1812|M.thermoresistible__bu YETIKGDASTGWMRLGAFLLAAAXRGARCARPTAPPCGTTTVQPGNPTRY
MAB_2724c|M.abscessus_ATCC_199 YETIRGDASVGWLRLGALLVLAAAFVGLVIN------------------R
MAV_4290|M.avium_104 YETVKGDASMAWMLLGAVVLAGAVLAGLLLN------------------R
***::**** .*: :* : * . .
Mb1517|M.bovis_AF2122/97 RLPT----------
Rv1481|M.tuberculosis_H37Rv RLPT----------
MLBr_01808|M.leprae_Br4923 RLPT----------
MMAR_2288|M.marinum_M RLPT----------
MUL_1490|M.ulcerans_Agy99 RLPT----------
Mflv_3659|M.gilvum_PYR-GCK RLPN----------
Mvan_2751|M.vanbaalenii_PYR-1 RLPN----------
MSMEG_3149|M.smegmatis_MC2_155 RLPG----------
TH_1812|M.thermoresistible__bu RAPGPAAWSCAPGR
MAB_2724c|M.abscessus_ATCC_199 RLPN----------
MAV_4290|M.avium_104 RLPA----------
* *